Join the Guix-Science community!
Guix channels let communities develop and maintain their own package collection at their own pace. As users of Guix in high-performance computing (HPC) and computational sciences, we have been developing several such channels. Those channels live under the Guix-Science umbrella, which recently moved to Codeberg. Over the last couple of months, we’ve been using this migration as an opportunity to strengthen scientific channels, both socially—by welcoming more contributions—and technically—by setting up infrastructure to improve the contribution and maintenance workflows.
In this post we look at the migration process, the available channels and what you can find there, and automation that is now in place. Our hope is that you will find it a good place to contribute your scientific packages.
Hello, Codeberg!
The Guix-Science umbrella was created several years ago, initially on GitHub. In September 2024, we, Guix-Science contributors, decided to give up GitHub and move to Codeberg. There are several reasons motivating this choice.
First, the code behind Codeberg (Forgejo) is free software; Codeberg itself is run by a non-profit that has proved to value transparency. In other words, it’s a whole new world compared to the walled-garden that GitHub is—one that is aligned with our convictions about open science and user autonomy. Forgejo supports a pull-request workflow so it’s not a significant change in that respect.
Another interesting and distinguishing aspect of Forgejo is the ongoing development of federation, a feature that will allow Forgejo instances to communicate together. This will support decentralization by reducing the incentives for users to all join the same instance—which is actually one of the recommendations in France’s report on forges in higher-education and research.
Like all modern forges, Forgejo has a rich HTTP programming interface. We wanted to take this migration as an opportunity to provide tight integration between the forge and our continuous integration tool.
Channels & packages
The Guix-Science umbrella now provides the following repositories:
- Guix-Science currently provides more than 700 packages in varying scientific domains: bioinformatics, statistics, HPC. Lately it received contributions in new domains: electronics design, physics, and neuroscience.
- Guix-Science-Nonfree is the channel we wish did not exist, providing proprietary scientific packages such as Geant4 and supporting tools such as CUDA.
- Guix-Past provides 130+ “packages from the past”, which can come handy when digging through old scientific code: Python 2, R versions as old as 2.15, Qt 4, old versions of Boost, etc.
- Guix-Jupyter, which is not a channel but instead an tool bringing reproducible deployment to Jupyter Notebook.
We are in the process of migrating the two remaining channels:
- Guix-CRAN, an automatic import of 20K packages from CRAN;
- Guix-Bioc, an automatic import of almost 9K packages from Bioconductor.
There is already a lot in there! If you maintain your own scientific package channel, we encourage you to join us and open pull requests to the relevant channel.
Automation
Packages in these channels are continuously built by the build farm at Inria. This is not only a quality assurance tool, but also a way to provide pre-built binaries for users.
Continuous integration and continuous delivery (CI/CD) is handled by Cuirass, a tool designed specifically to support Guix. In recent months, we started adding Forgejo support to Cuirass. The first step was to allow Cuirass to create new jobsets in response to webhooks corresponding to a new pull request, and to maintain metadata linking those jobsets to the pull request on Codeberg.
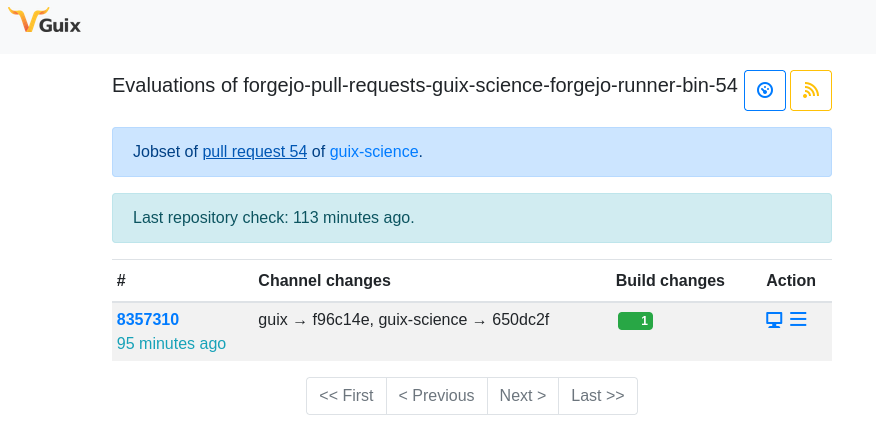
Technically, we added per-project webhooks on Codeberg so that it would
send POST requests to
https://guix.bordeaux.inria.fr/admin/forgejo/event, and we
configured
the nginx server for guix.bordeaux.inria.fr to require so-called basic
authentication for that endpoint.
We are now in the process of implementing communication in the other direction: allowing Cuirass to report success or failure to Forgejo by leaving messages in the corresponding pull request. This will provide an experience similar to what developers have come to enjoy with tools like GitLab-CI, specifically tailored to the needs of Guix.
In addition to CI/CD, we devised
teams that are
automatically notified when a pull request touching their domain comes
in—the mapping from teams to files is specified in CODEOWNERS files.
This is another nifty tool that we hope will help reduce the chances
that pull requests go unnoticed.
We need you! 🫵
All this technical work is really in support of one goal: bringing the scientific community together around tools that enable reproducible research and open science.
If you already have your own channel for scientific packages in your domain, consider contributing them to Guix-Science for the benefit of the broader community; in return, you get peer review, continuous integration, pre-built binaries, and new friendly co-workers. We recommend starting with small bites, preferably one package at a time, to make it easy on reviewers and on you.
If you’re into bioinformatics, HPC, physics, electronics design, neuroscience, or any other domain that may be represented in Guix-Science, consider creating or joining a team to help review packaging work in those areas.
Join us in building a commons for the scientific community!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).






