Guix-HPC Activity Report, 2021
This document is also available as PDF (printable booklet).
Guix-HPC is a collaborative effort to bring reproducible software deployment to scientific workflows and high-performance computing (HPC). Guix-HPC builds upon the GNU Guix software deployment tools and aims to make them useful for HPC practitioners and scientists concerned with dependency graph control and customization and, uniquely, reproducible research.
Guix-HPC was launched in September 2017 as a joint software development project involving three research institutes: Inria, the Max Delbrück Center for Molecular Medicine (MDC), and the Utrecht Bioinformatics Center (UBC). GNU Guix for HPC and reproducible science has received contributions from additional individuals and organizations, including CNRS, the University of Paris (Diderot), the University of Tennessee Health Science Center (UTHSC), the Leibniz Institute for Psychology (ZPID), Cray, Inc. (now HPE), and Tourbillion Technology.
This report highlights key achievements of Guix-HPC between our
previous
report
a year ago and today, February 2022. This year was marked by exciting
developments for HPC and reproducible workflows: the release of
GNU Guix 1.3.0 in
May, the
ability to tune packages for a CPU micro-architecture with the --tune
option, improved Software Heritage support, new releases of Guix-Jupyter
and the Guix Workflow Language (GWL), support for POWER9 CPUs and
on-going work porting to RISC-V, and more.
Outline
Guix-HPC aims to tackle the following high-level objectives:
- Reproducible scientific workflows. Improve the GNU Guix tool set to better support reproducible scientific workflows and to simplify sharing and publication of software environments.
- Cluster usage. Streamlining Guix deployment on HPC clusters, and providing interoperability with clusters not running Guix.
- Outreach & user support. Reaching out to the HPC and scientific research communities and organizing training sessions.
The following sections detail work that has been carried out in each of these areas.
Reproducible Scientific Workflows
Supporting reproducible research workflows is a major goal for Guix-HPC. The ability to reproduce and inspect computational experiments—today’s lab notebooks—is key to establishing a rigorous scientific method. UNESCO’s Recommendation on Open Science, published in November 2021, recognizes the importance of free software in research and further notes (§7d):
In the context of open science, when open source code is a component of a research process, enabling reuse and replication generally requires that it be accompanied with open data and open specifications of the environment required to compile and run it.
This key point is often overlooked: the ability to reproduce and inspect the software environments of experiments is a prerequisite for transparent and reproducible research workflows.
To that end, we work not only on deployment issues, but also upstream—ensuring source code is archived at Software Heritage—and downstream—devising tools and workflows for scientists to use. The sections below summarize the progress made on these fronts and include experience reports by two PhD candidates showing in concrete terms how Guix fits in reproducible HPC workflows.
Workflow Languages
The Guix Workflow Language (or GWL) is a scientific computing extension to GNU Guix's declarative language for package management. It allows for the declaration of scientific workflows, which will always run in reproducible environments that GNU Guix automatically prepares. In the past year the GWL has received several bug fixes and infrastructure for detailed logging; it also gained a DRMAA process engine to submit generated jobs to any HPC scheduler with an implementation of DRMAA, such as Slurm and Grid Engine. This was made possible through the newly released high-level Guile bindings to DRMAA version 1. We released version 0.4.0 of the GWL on January 29.
Earlier in January, we announced ccwl, the Concise Common Workflow Language. ccwl is a workflow language with a concise syntax compiling to the Common Workflow Language (CWL). While GWL offers a novel workflow language with integrated deployment via Guix, ccwl instead aims to leverage tooling around the popular Common Workflow Language while addressing some of its limitations. We published a detailed article introducing ccwl and expounding its merits. ccwl significantly cuts short on the verbosity of CWL, thus removing one of the barriers to its wider adoption. ccwl is implemented as a domain specific language embedded in GNU Guile, and interoperates with GNU Guix to provide reproducibility. ccwl also aims to minimize frustration for users by providing strong compile-time error checking and high-quality error messages. We also plan to pre-package commonly used command-line scientific tools into ready-made ccwl workflows. Work on these exciting new features is already underway.
Reproducible Software Deployment for Jupyter
We announced Guix-Jupyter two years ago, with two goals: making notebooks self-contained or “deployment-aware” so that they automatically deploy the software (and data!) that they need, and making said deployment bit-reproducible. Earlier this year, we published version 0.2.2 as a bug-fix release.

Guix-Jupyter is implemented as a Jupyter kernel: it acts as a proxy between the notebook and the programming language notebook cells are written in. It interprets annotations found in the notebook to deploy precisely the right software packages needed to run the notebook. We believe this is a robust approach to address the Achilles’ heel that software deployment represents for reproducible computations with Jupyter.
Yet, because Binder and its associated services
and tools are a popular way to deploy Jupyter notebooks, we wanted to
offer an alternative solution integrated with Binder. Under the hood,
Binder builds upon
repo2docker, a tool to
build Docker images straight from source code repositories. Repo2docker
has a number of back-ends called buildpacks to handle packaging
metadata in a variety of formats: when a setup.py file is available,
software is deployed using standard Python tools, the presence of an
install.R file leads to deployment using GNU R, an apt.txt file
instructs it to install software using Debian’s package manager, and so
on.
As part of a three-month internship at Inria, Hugo Lecomte implemented a
Guix buildpack for repo2docker. If a guix.scm or a manifest.scm
file is found in the source repository, repo2docker uses it to populate
the Docker image being built. Additionally—and this is a significant
difference compared to other buildpacks—, software deployed with Guix
can be pinned at a specific revision: if a
channels.scm file is
found, the buildpack passes it to guix time-machine; this ensures that
software is deployed from the exact Guix revision specified in
channels.scm.
This Guix buildpack for repo2docker has been submitted upstream and reviewed, but as of this writing it has yet to be merged. We believe it provides another convenient way for Jupyter Notebook users to ensure their code runs in the right software environment.
Ensuring Source Code Availability
Guix lets users re-deploy software environments, for instance via
guix time-machine.
This is possible because Guix can rebuild software, which, in turn, is
only possible if source code is permanently available. Since
2019
Guix developers collaborate with Software Heritage (SWH) to make that a
reality. A lot has been achieved since then but some challenges remained before we
could be sure that SWH would archive every piece of source code Guix
packages refer to.
One of the main roadblocks we identified early
on
are source code archives—
tar.gz and similar files, colloquially known
as “tarballs”. SWH, rationally, stores the contents of these archives,
but it does not store the archives themselves. Yet, most Guix package
definitions refer to tarballs; Guix expects to be able to download those
tarballs and to verify that they match.
How do we deal with this impedance mismatch?
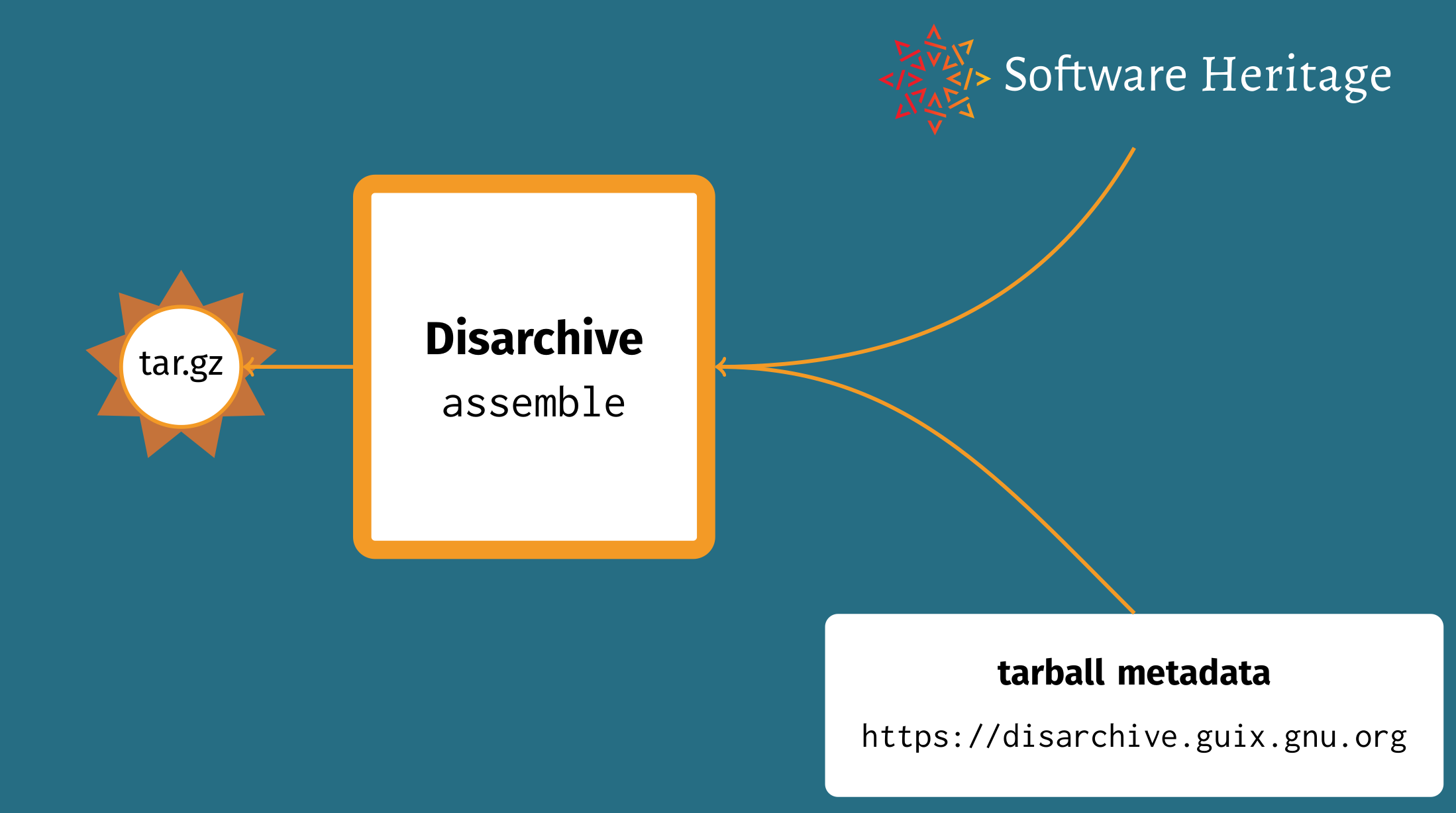
Last year, Guix developer Timothy Sample had just started work to address this. Timothy developed a tool called Disarchive that supports two operations: “disassembling” and “reassembling” tarballs. In the former case, it extracts tar and compression metadata along with an identifier (SWHID) pointing to contents available at SWH; in the latter case, Disarchive assembles content and metadata to recreate the tarball as it initially existed. From there we create a Disarchive database that maps cryptographic hashes of tarballs to their metadata.
This year we deployed, on the Guix build farm, infrastructure to
continuously build the
database and to publish it
at disarchive.guix.gnu.org. We
added support in Guix so that it can use Disarchive + SWH as a fallback
when downloading a tarball from its original URL fails, significantly
improving source code archival coverage.
Beyond Guix, this work is crucial for all the deployment tools that rely
on the availability of tarballs—Brew, Gentoo, Nix, Spack, and other
package managers, but also scientific workflow tools such as
Maneage and individual Dockerfiles and
scripts. This led SWH and the Sloan Foundation to allocate a
grant
so that Timothy Sample could address some of the remaining challenges.
Among those, Timothy has already been able to expand Disarchive compression support beyond gzip—version 0.4.0 adds support for xz, the second most popular compression format for tarballs. To have a clear vision of the progress being made, Timothy has been publishing periodic Preservation of Guix Reports. The latest one shows that archival coverage for all the Guix revisions since version 1.0.0 is at 72%; the breakdown by revision shows that coverage reaches 86% for recent commits. Simon Tournier has been carefully monitoring coverage and discussing with other Guix developers and with the SWH team to identify reasons why specific pieces of source code would not be archived. Ludovic Courtès had the pleasure to join the SWH Fifth Anniversary event, on behalf of the Guix team, to show all the progress made and to discuss the road ahead.
Tuning Packages for a CPU
GNU Guix is now well known for supporting “reproducibility”, which is really twofold: it is first the ability to re-deploy the same software stack on another machine or at a different point in time, and second the ability to verify that binaries being run match the source code—the latter is what reproducible builds are concerned with.
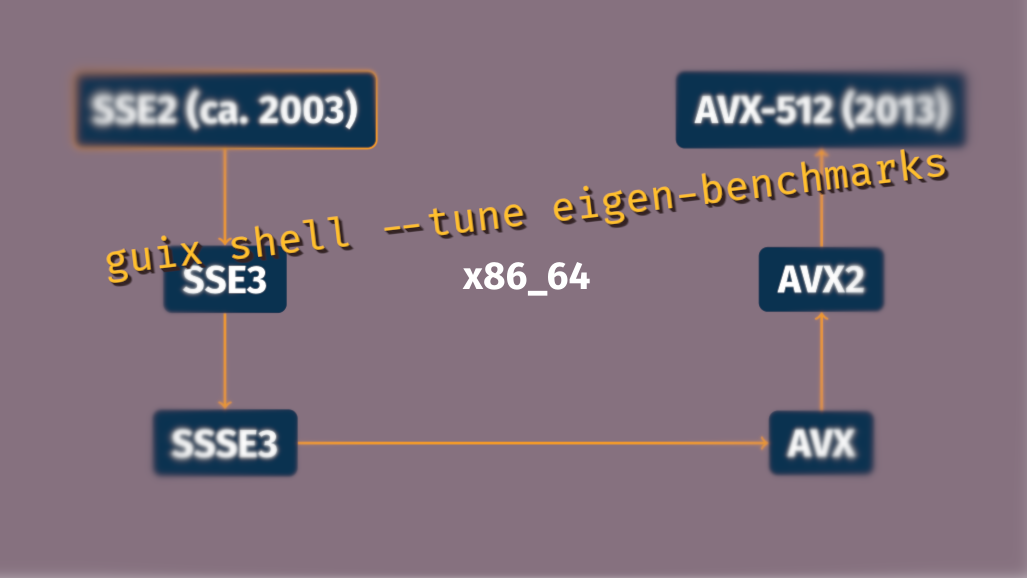
However, in HPC circles there is the entrenched perception that reproducibility is antithetic to performance. Practitioners are especially concerned with the performance of the Message Passing Interface (MPI) implementations on high-speed network devices, and with the ability of code to use single-instruction/multiple-data (SIMD) extensions of the latest CPUs—such as AVX-512 on x86_64, or NEON on ARMv8. We showed that these concerns are largely unfounded in a 2018 article on achieving performance with portable binaries and in a 2019 article on Open MPI.
The former article showed how performance-sensitive C code is already taking advantage of function multi-versioning (FMV). There remain cases, though, where this technique is not applicable. As a result, GNU/Linux distributions—from Guix to Debian and CentOS—that distribute binaries built for the baseline x86_64 architecture miss out on SIMD optimizations. A notorious example of packages that do not support FMV is C++ header-only libraries, such as the Eigen linear algebra library.
To address this, we introduced what we call package
multi-versioning:
with the new --tune package transformation option, Guix users can
obtain a package variant specifically tailored for the host CPU. Yet,
users can avoid rebuilding time-consuming local builds if a pre-built
binary for the same CPU variant is available on-line.
While building a package with -march=native (instructing the compiler
to optimize for the CPU of the build machine) leaves no trace, using
Guix’s --tune is properly recorded in metadata. For example, a Docker
image built with guix pack --tune --save-provenance contains, in its
metadata, the CPU type for which it was tuned, allowing for independent
verification of its binaries. This is to our knowledge the first
implementation of CPU tuning that does not sacrifice reproducibility.
Packaging
The package collection that comes with Guix keeps growing. It now contains more than 20,000 curated packages, including many scientific packages ranging from run-time support software such as implementations of the Message Passing Interface (MPI), to linear algebra software, to statistics and bioinformatics modules for R.
The Julia programming language has been gaining traction in the
scientific community and efforts in Guix reflect that momentum. At the time of the previous report,
February 2021, Guix included a dozen Julia packages. Today, January 2022,
it includes more than 260 Julia packages, from bioinformatics software
such as BioSequence.jl to
machine learning software like Zygote.jl. Under the hood, the Julia build
system in Guix has been
improved; in particular, it now supports both parallel builds and
parallel tests, providing a significant speedup. It also
allows the built-in Julia package manager Pkg to find packages
already installed by Guix.
In 2021, we added the popular PyTorch machine learning framework to our
package collection. While it had long been available via pip, the
Python package manager, we highlighted in a blog
post things that we as users do not
notice about packages: what is inside of them, and the work behind it.
We showed that the requirements for Guix packages to build software from
source and to avoid bundling external dependencies are key to
transparency, auditability, and provenance tracking—all of which are
ultimately the foundations of reproducible research.
Many scientific packages were upgraded: the Dune finite element libraries have been updated to 2.7.1, the Python bindings to Gmsh were updated to 7.1.11, PETSc and related packages were updated to 3.16.1, to name a few. Run-time support packages such as MPI libraries also received a number of updates.
Statistical and bioinformatics packages for the R programming language have seen regular comprehensive upgrades, closely following updates to the popular CRAN and Bioconductor repositories. At the time of this writing Guix provides a collection of more than 1900 reproducibly built R packages, making R one of the best supported programming environments in Guix.
Core packages have seen important changes; in particular, packages are
now built with GCC 10.3 by default (instead of 7.5), using the GNU C
Library version 2.33. The style of package inputs has been
considerably
simplified; together
with the introduction of guix style
for automatic formatting, we hope it will make it easier to get started
writing new packages.
Supporting POWER9 and RISC-V CPUs
In April 2021, Guix gained support for POWER9 CPUs, a platform that some HPC clusters build upon. While support in Guix—and in the broader free software stack—is not yet on par with that of x86_64, it is gradually improving. The project’s build farm now has two beefy POWER9 build machines.
While it is perhaps early days to call RISC-V an HPC platform, there are indicators that this may happen in the near future with investments from the USA, the EU, India, and China.
Together with Chris Batten of Cornell and Michael Taylor of the University of Washington, Erik Garrison and Pjotr Prins are UTHSC PIs responsible for creating a new NSF-funded RISC-V supercomputer for pangenomics. It will incorporate GNU Guix and the GNU Mes bootstrap, with input from Arun Isaac, Efraim Flashner and others. NLNet is also funding the GNU Mes RISC-V bootstrap project with Ekaitz Zarraga and Jan Nieuwenhuizen. We aim to continue adding RISC-V support to GNU Guix at a rapid pace.
Why is the combination of GNU Mes and GNU Guix exciting for RISC-V? First of all, RISC-V is a very modern modular open hardware architecture that provides further guarantees of transparency and security. It extends reproducibility to the transistor level and for that reason generates interest from the Bitcoin community, for example. Because there are no licensing fees involved, RISC-V is already a major force in IoT and will increasingly penetrate hardware solutions, such as storage microcontrollers and network devices, going all the way to GPU-style parallel computing and many-core solutions with thousands of cores on a single die. GNU Mes and GNU Guix are particularly suitable for RISC-V because Guix can optimize generated code for different RISC-V targets and is able to parameterize deployed software packages for included/excluded RISC-V modules.
On the way to a reproducible PhD thesis
GNU Guix and Org mode form a powerful association when it comes to setting up a PhD thesis workflow. On one hand, GNU Guix allows us to ensure an experimental software environment is reproducible across various high-performance testbeds. On the other hand, we can take advantage of the literate programming paradigm using Org mode to describe the experimental environment as well as the experiments themselves, then post-process and reuse the results in final scientific publications.
The ongoing work of Marek Felšöci at Inria is an actual attempt for a reproducible PhD thesis relying on the conjunction of GNU Guix and Org mode. The thesis project resides in a Git repository where a dedicated Org file describes and explains all of the source code and procedures involved in the construction of the experimental software environment, the execution of experiments as well as the gathering and the post-processing of the results. This includes a Guix channel file, scripts for running the experiments, parsing the output logs, producing figures and so on.
Other Org documents of the repository may then build on these results and produce the final publications, such as research reports, articles and slideshows, in various formats. As an existing publication example we can cite the research report #9412 and the associated technical report #0513 providing a literate description of the environment and the experiments the study presented in the research report relies on.
In the end, the entire process of setting up the software environment, running experiments, post-processing results and publishing documents is automated using continuous integration.
The result of the continuous integration is publicly available as a collection of web pages and PDF documents hosted using GitLab Pages.
The initiative does not stop here. There is an effort to transform this monolithic setup into independent modules with the aim to share and reuse portions of the setup in other projects within the research team.
Feedback from using Guix to ensure reproducible HPC experiments
Philippe Swartvagher (Inria) took the opportunity of writing an article on the impact of execution tracing on complex HPC applications to discover how GNU Guix could help perform reproducible experiments. The article studies the impact of tracing on application performance, evaluates solutions to reduce this impact, and explores clock synchronization issues when distributed applications are traced. The paper is still under review.
The software stack considered in the article is made of several libraries (StarPU, Chameleon, PM2 and FxT), all of them being already packaged in GNU Guix, in the Guix-HPC channel. Manually installing this software stack can be painful, the set of compilation options is wide and desired options can change from an experience to another, to see their impact. Correctly compiling the software stack before each experiment and tracking its current state can be pretty tedious.
This source of headaches disappears with GNU Guix, especially with the help of
package
transformations.
For instance, --with-input allowed us to use of PM2 instead of the Open MPI
as the communication engine, --with-commit was handy to select a
specific commit of a library (for instance to compare performance before
and after a specific change), and --with-patch was
convenient to apply code modification for a specific experiments (for
instance modifications not suited to be included upstream, but required for the
experiment). These package tranformations, used with guix environment (the
predecessor of guix shell),
removes the burden of compiling the correct version of each software before
each experiment.
This intense use of package transformations lead to some corner cases of GNU Guix features and raises several issues.
To ensure reproducibility of experiments made with GNU Guix, software versions
have to be pinned and saved along with scripts to launch the experiments.
guix describe
and guix time-machine
are the two GNU Guix's commands to pin revisions and execute
applications built from these precise revisions. Making the experimental
scripts publicly
available is
another step to achieve a reproducible article. It requires us to clearly
organize experiments, describe their goals and workings and ensure the maximum
independence from cluster specificities (or document which changes are
necessary to launch the experiments on another cluster). When the repository
describing the experiments is completed, archiving it on Software Heritage and
providing the obtained ID in the paper to easily retrieve the scripts is
effortless.
This first paper with GNU Guix was a great opportunity to discover the help
provided by GNU Guix, its ecosystem and support. It also showed areas where
documentation can be improved regarding the workflow to ensure reproducibility
of the experiments—from using guix describe to pin versions, to obtaining
an ID to easily cite the scripts in a paper. Moreover, there are still pending
questions about the best way to generalize experimentation scripts and make them
independent from the clusters being used—e.g., how to deal with different
job schedulers, file systems, and how to provide
instructions to replicate experiments even without Guix.
Cluster Usage and Deployment
At UTHSC, Memphis (USA), we are running an 11-node large-memory HPC Octopus cluster (264 cores) dedicated to pangenome and genetics research. In 2021 more SSDs and RAM were added. Notable about this HPC is that it is administered by the users themselves. Thanks to GNU Guix we install, run and manage the cluster as researchers (and roll back in case of a mistake). UTHSC IT manages the infrastructure, i.e., physical placement, routers and firewalls, but beyond that there are no demands on IT. Thanks to out-of-band access we can completely (re)install machines remotely. Octopus runs GNU Guix on top of a minimal Debian install and we are experimenting with pure GNU Guix nodes that can be run on demand. Lizardfs is used for distributed network storage. Almost all deployed software has been packaged in GNU Guix and can be installed by regular users on the cluster without root access.
At GLiCID (Nantes, France) we are in the process of merging two existing
HPC clusters (10,000+ cores). The first cluster (based on Slurm +
CentOS) already offers the guix command to our users as well as some
specific software on our own Guix channel, since a few years. This
merger involves a lot of change, including identity management. We
wanted to take advantage of this profound change to be more ambitious
and explore automated generation of part of the core infrastructure,
using virtual machines generated by guix system, deployed on
KVM+Ceph. We aim to eventually replace as many of these deployed machines
as possible, adjusting Guix system services and implementing new ones
as we go, benefiting the wider community.
Outreach and User Support
Articles
The following articles were published in the November 2021 edition of 1024, the magazine of the Société Informatique de France (SIF), the French computer science society:
- Konrad Hinsen, La Reproductibilité des calculs coûteux
- Ludovic Courtès, Reproduire les environnements logiciels : un maillon incontournable de la recherche reproductible
This article appeared in the March 2021 special edition of French-speaking GNU/Linux Magazine France:
- Ludovic Courtès, Déploiements reproductibles dans le temps avec GNU Guix
The following article introducing the most recent addition to the PiGx framework of reproducible workflows backed by Guix is awaiting peer-review and has been submitted to the medRxiv preprint server:
- Vic-Fabienne Schumann et al., COVID-19 infection dynamics revealed by SARS-CoV-2 wastewater sequencing analysis and deconvolution
Talks
Since last year, we gave the following talks at the following venues:
- JCAD conference, Dec. 2021 (Ludovic Courtès)
- Software Heritage Firth Anniversary, joint event with UNESCO, Nov. 2021 (Ludovic Courtès)
- TREX Build System Hackathon, Nov. 2021 (Ludovic Courtès)
- PackagingCon, Nov. 2021 (Ludovic Courtès)
- “Pour une recherche reproductible”, MAiMoSIne, SARI, GRICAD, Nov. 2021 (P.-A. Bouttier)
- RDA 18th plenary, Software Source Code, Nov. 2021 (P.-A. Bouttier)
- Reproducible FAIR+ Workflows and the CCWL, at the US NIH National Cancer Institute in Oct. 2021 (Pjotr Prins, Arun Isaac)
- special event on reproducibility of the Société Informatique de France (French computer science society), May 2021 (Konrad Hinsen, Ludovic Courtès)
We also organised the following events:
- the first on-line workshop on the reproducibility of software environments for French-speaking scientists, engineers, and system administrators, on May 17–18th, 2021 with up to 80 participants.
- “Declarative and minimalistic computing” track at FOSDEM
Training Sessions
A training session on computational reproducibility for high-energy sessions took place at the Centre de Physique des Particules de Marseille in April/May 2021. It included a hands-on session about Guix.
For the French HPC Guix community, we have set up a monthly on-line event called “Café Guix”, started in October 2021. Each month, a user or developer informally presents a Guix feature or workflow and answers questions.
Personnel
GNU Guix is a collaborative effort, receiving contributions from more than 90 people every month—a 50% increase compared to last year. As part of Guix-HPC, participating institutions have dedicated work hours to the project, which we summarize here.
- Inria: 2 person-years (Ludovic Courtès and the contributors to the Guix-HPC channel: Emmanuel Agullo, Marek Felšöci, Nathalie Furmento, Hugo Lecomte, Gilles Marait, Florent Pruvost, Matthieu Simonin, Philippe Swartvagher)
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC): 2 person-years (Ricardo Wurmus and Mădălin Ionel Patrașcu)
- University of Tennessee Health Science Center (UTHSC): 3+ person-years (Efraim Flashner, Bonface Munyoki, Fred Muriithi, Arun Isaac, Jorge Gomez, Erik Garrison and Pjotr Prins)
- Utrecht Bioinformatics Center (UBC): 0.1 person-year (Roel Janssen)
- University of Paris (Diderot): 0.5 person-year (Simon Tournier)
Perspectives
Guix availability on scientific computing clusters remains a top priority. More HPC practitioners—researchers, engineers, and system administrators—are adopting Guix and showing interests, from reproducible research to flexible deployment of virtual machines. We expect to continue to work on these two complementary fronts: streamlining the use of reproducible packs, and reaching out to system administrators and cluster users, notably through training sessions.
Upstream, we will continue to work with Software Heritage with the goal of achieving complete archive coverage of the source code Guix refers to. We have identified challenges related to source code availability; this will probably be one of the main efforts in this area for the coming year.
Downstream, a lot of work has happened in the area of reproducible research tools. Our package collection has grown to include more and more scientific tools. Tools like the Guix Workflow Language and Guix-Jupyter have matured; along with the PsychNotebook service, they bridge the gap between reproducible software deployment and reproducible scientific tools and workflows. We also showed how to achieve high performance while preserving provenance tracking, which we hope dispels the entrenched perception in HPC circles that reproducibility and performance are antithetic.
Our work happens in a context of growing awareness of the importance of software and software environments in research workflows. UNESCO’s Recommendation on Open Science and, for example, the Second French Plan for Open Science are two illustrations of that.
We gave demonstrations of what Guix brings to scientific workflows and we expect to continue to show that reproducible scientific workflows are indeed a possibility. Working on the tools and workflows directly in the hands of scientists will be a major focus of the coming year. We want to contribute to raising the bar of what scientists come to expect in terms of reproducible workflows.
There’s a lot we can do and we’d love to hear your ideas!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).








