Guix-HPC Activity Report, 2025

This document is also available as PDF (printable booklet)
Guix-HPC is a collaborative effort to bring reproducible software deployment to scientific workflows and high-performance computing (HPC). Guix-HPC builds upon the GNU Guix software deployment tools and aims to make them useful for HPC practitioners and scientists concerned with dependency graph control and customization and, uniquely, reproducible research.
Guix-HPC started as a joint software development project involving three research institutes: Inria, the Max Delbrück Center for Molecular Medicine (MDC), and the Utrecht Bioinformatics Center (UBC). Guix for HPC and reproducible research has since received contributions from many individuals and organizations, including CNRS, Université Paris Cité, the University of Tennessee Health Science Center (UTHSC), Cornell University, and AMD. HPC remains a conservative domain but over the years, we have reached out to many organizations and people who share our goal of improving upon the status quo when it comes to software deployment.
This report—our eighth report!—highlights key achievements of Guix-HPC between our previous report a year ago and today, February 2026. This year was marked by exciting developments for HPC and reproducible workflows. Significant advances were made in integrating Guix into the complex software landscape of HPC, taking the roles of software manager, workflow execution engine, backend for generating container images, or provider for the complete operating system layer. This year was also marked by the migration to Codeberg, a non-profit and community-led European platform whose main objective is to facilitate the expansion of the contributor community. Finally, support for reproducing computations from the past was also much improved. And, as usual, we have been using Guix for research, and teaching other researchers how to get started.
Outline
Guix-HPC aims to tackle the following high-level objectives:
- Reproducible scientific workflows. Improving the GNU Guix tool set to better support reproducible scientific workflows and to simplify the sharing and publication of software environments.
- Cluster usage. Streamlining Guix deployment on HPC clusters, and providing interoperability with clusters not running Guix.
- Outreach & user support. Reaching out to the HPC and scientific research communities and organizing training sessions.
The following sections provide details of the work carried out in each of these areas.
Reproducible Scientific Workflows
Supporting reproducible research workflows is a major goal for Guix-HPC. This section looks at progress made on packaging and tooling.
Packaging
Work on HPC and scientific packages happens both in Guix proper and in the channels maintained by Guix-Science.
Strengthened HPC and Scientific Packages in Guix
Foundational HPC and scientific packages are maintained as part of the the more than 30,000 packages available in Guix itself. Quite a few key packages were added, such as Kokkos and related libraries, oneTBB, and oneDNN. Many key packages were upgraded, including Open MPI and supporting libraries, PETSc and SLEPc, OpenBLAS, PyTorch and its many dependencies, GROMACS, OpenCascade, and many more.
Development work in Guix is organized around autonomous teams. This year the Science team was complemented by an HPC team—the latter being responsible for the lower-level HPC support packages. With the switch to Codeberg as the development platform, pull requests are automatically assigned and labeled for the relevant team.
Guix-Science Collaboration
The Guix-Science project aims at providing a comprehensive, community-driven, scientific software catalog suitable for the global scientific community, with a particular attention to the needs of supercomputer administrators. One of the benefits is to share the packaging effort, particularly maintenance.
More than 200 packages were added to Guix-Science this year, including MPI implementations (e.g., MVAPICH), runtime support libraries (e.g., PaRSEC) or applications (e.g., PLUMED, Quantum ESPRESSO). Moreover, core components or full stack framework of digital health applications were added as part of an effort to the French Digital Health Research Program (PEPR Santé Numérique).
Many packages that were previously in the Guix-HPC channel have been migrated to Guix-Science and consolidated, including StarPU, Chameleon, PaStiX, Composyx, qr_mumps, Netlib HPL, Gyselalib++ and PDI. We aim at making Guix-Science the reference channel for scientific software and lower the barrier to entry for scientists willing to contribute.
Continuous Integration
Cuirass is the continuous integration tool running on the Guix-HPC build farm.
Cuirass has been extended to support pull request evaluation from GitLab and Forgejo instances. It now evaluates all the Guix-Science pull requests, builds the modified packages and sends results back to the forge instance.
An additional quality assurance pipeline based on Forgejo Action
has been set up in order to automate quality assurance of packages
submitted to the Guix-Science channels. It currently uses a custom
guix diff command (source code is available) to
list the new/updated packages and runs guix lint on them.
Concise Common Workflow Language (ccwl)
The Concise Common Workflow Language
(ccwl)
is a concise syntax to express Common Workflow Language
(CWL) workflows. It is implemented as an
EDSL (Embedded Domain Specific Language) in Guile Scheme. Unlike
workflow languages such as the Guix Workflow Language (GWL), ccwl is
agnostic to deployment. It does not use Guix internally to deploy
applications. It merely picks up applications from PATH and thus
interoperates well with Guix and any other package manager of the
user's choice. ccwl also compiles to CWL and thus reuses all
tooling built to run CWL workflows. Workflows written in ccwl may be
freely reused by CWL users without impediment, thus ensuring smooth
collaboration between ccwl and CWL users.
ccwl 0.5.0 was released in January 2026. ccwl 0.5.0 adds support for separated prefix arguments. As with previous versions, ccwl 0.5.0 comes with many practical examples in the manual.
ravanan
ravanan is a new Common Workflow Language (CWL) implementation that is powered by Guix and provides strong reproducibility guarantees. ravanan runs CWL workflows as jobs on a HPC batch system. ravanan uses Guix to manage all packages used by steps in the workflow. In addition, it takes inspiration from Guix and uses a content-addressed store to provide strong caching of intermediate results. Thanks to these, ravanan never runs the same computation twice and its cache never goes stale.
Other salient features of ravanan include:
- One-to-one correspondence between steps in the CWL workflow and jobs on the HPC batch system;
- Clear logging for every job run;
- Jobs never write directly to the shared network filesystem on HPC, which is essential for good performance;
- Jobs never write to
/tmp.
Lack of reproducibility and increasingly complex software are a growing concern in scientific workflows. Many of these challenges have been effectively addressed by Guix. ravanan takes this to the next level by seamlessly integrating Guix with a well-established standards-based workflow language like CWL.
ravanan had its second release—version 0.2.0—in November 2025.
Reproducible Research in Practice
In this section, we look at the variety of ways Guix is used to support reproducible research work.
A Guide for Reproducible Research
We consolidated a guide on using Guix for reproducible research workflows in the Guix Cookbook, based on an earlier blog post. This is a hands-on guide targeting scientists, which details 4 steps to reproducible computational workflows:
- setting up the environment;
- recording the environment;
- ensuring long-term source code archiving with Software Heritage;
- referencing the software environment.
The guide has already proved helpful in getting new people started with Guix for their research work.
Deploying Linear Solvers in an Industrial Context
In 2025, Esragul Korkmaz (of the Concace research team at Inria) wrote an internal technical report comparing 26 different solvers for large coupled sparse/dense linear systems arising from aeroacoustic simulations at Airbus. This research was conducted in collaboration with Inria and Airbus, within the framework of the Mambo project (funded by DGAC). Crucially, the report's appendix details a robust methodology for achieving reproducibility, with Guix utilization as the core component. This approach was necessary because the sheer quantity and complexity of the dependencies for the 26 solvers would have otherwise rendered reliable reproducibility unmanageable. This work, thus, provides a powerful testimonial for achieving reproducible computational workflows, even within demanding and complex HPC environments. She is now working on a research report that will be made publicly available.
Guix in Digital Health
Digital Health is highly quantitative and relies on processing a wide range of data. The optimization of diagnostic or therapeutic strategies as personalized routines or new specialized drugs, their development and validation, rest on operating and integrating large volumes of multi-scale heterogeneous health data. Hence, it needs to run multi-step pipelines (workflows) where each step implies different software. In addition, some of these software packages might be considered as medical device, therefore patient safety requires a clear identification and tracking of all the components. It means each source code must be intrinsically identified and the associated machine-readable binary artefact must be inspectable.
Because effective digital health requires the combination of such software packages with data and runtime environments to produce analyses and get results eventually, Guix provides by design clear source code identifications and inspectable binaries. Thus, Guix helps in formalizing, building, and deploying reproducible computational environments for research in digital health. Moreover, research in digital health suffers the same replication and reproducibility defect as most of the other research fields, where computational environments constitute one source of variability, mainly coming from the insufficient capture of their details.
This year, the effort has been continued and strengthened. Core components or complete applications required by neuroimaging has been merged to Guix-Science channel (see dedicated section above) and are quickly available to scientific practitioners. Moreover, talks, training and hands-on sessions have allowed to federate broader multidisciplinary research community active in digital health. We pursue the aim at making Guix a robust alternative framework for deploying reproducible computational environments required by digital health research.
Guix in Medical Physics and Nuclear Medicine
A team at South Australia Medical Imaging is developing Spider, a software pipeline for performing image-based radiation dosimetry for radionuclide therapy patients. Producing dose estimates involves various tasks including image conversion, image registration, organ and lesion segmentation, and potentially Monte Carlo radiation track structure simulations. Spider is in the early stages of development, but it aims to glue together existing free software that specialises in each of these tasks.
The reproducibility guarantees afforded by Guix provide the foundation for long-term development and maintenance of the pipeline amidst an ever-evolving software landscape. An important prerequisite is to write Guix package definitions for software to be included in the pipeline. Thanks in particular to contributions by Ghislain Vaillant and Jake Forster, the Guix proper, Guix-Science, and Guix-Science-Nonfree channels together have many programs and libraries that are useful for radionuclide therapy dosimetry, including dcm2niix, elastix, Geant4, EGSnrc, and STIR, as well as up-to-date versions of ITK and ITK-SNAP.
The pipeline is free software and we aim to provide a legitimate alternative to proprietary solutions that are prohibitively expensive. Ultimately, success will be measured by whether it gains approval for use as a software-based medical device, and we think using Guix as a deployment platform will strengthen our application.
Guix in Biology
Le Souchu et al. utilized Guix for the article Intra- and interspecific variations in flight performance of oak-associated Agrilinae (Coleoptera: Buprestidae) using computerised flight mills as a tool to ensure long-term reproducibility of data analyses performed with R, providing the associated recipe and documentation in an archive and a Git repository hosted on governmental and institutional servers. Although some authors had used Guix before, this was their first published work employing it, marking their commitment to more reproducible research in future projects and better incorporating these practices into student training in their field. The article was published in Peer Community Journal, a diamond open-access journal with open and reproducible science at its core.
Guix in Material Science
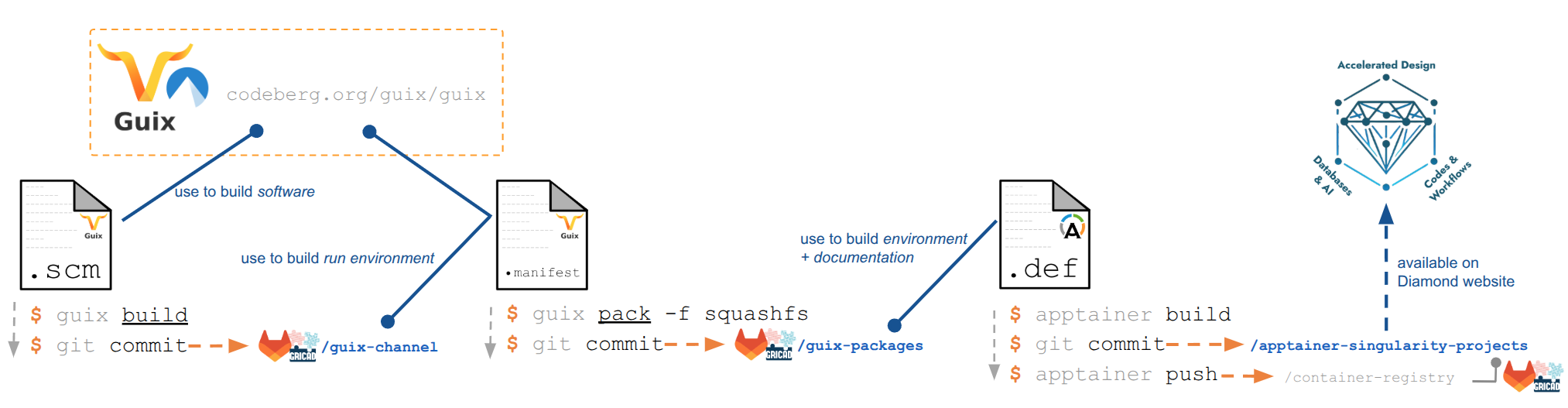
The DIAMOND numerical platform of the DIADEM research program, at a slower rate, kept expanding its material science related channels.
In the previous report, a continuous integration pipeline has been presented. The pipeline uses the DIAMOND Guix channel as a source to first create SquashFS archives, using guix pack. The latter are then used as a base image to build a fully reproducible Apptainer containers, embedding the complete runtime environment and documentation to use the container on HPC infrastructure.
The end goal of those Guix-based Apptainer images is to be used in numerical workflows. With this being done, a platform paper has been submitted in December 2025, Reproducible container solutions for codes and workflows in materials science. It presents the method, applications, use cases, and benchmarks showing negligible overhead for Guix-backed containers, compared to machine-compiled code. Another milestone regarding the DIAMOND platform is the first package supporting GPU offloading using CUDA libraries.
Efforts will now be aimed at contributing to Guix's main channels to provide material science packages outside of the DIADEM community, as well as improving GPU support for said packages.
Application Profiling and Performance Analysis
The HPC development practice goes in cycles. We write code, we analyze the performances of the program, we optimize the code, and we measure the performances once again (repeat one or more times). This process requires tools for HPC software profiling and performance analysis.
Once we have a free and reproducible software environment, it would be a pity to quit it for finally analyzing the performances of our program. Beside the already available packages for LIKWID, PAPI, GNU Gprof, GNU Gprofng, and the Valgrind suite (just to cite some), new packages for the Score-P measurement infrastructure, the Scalasca parallel profiler, and the Cube performance visualization tool have been contributed to the Guix channel for a richer HPC development environment.
Cluster Usage and Deployment
The sections below highlight the experience of cluster administration teams and report on tooling developed around Guix for users and administrators on HPC clusters.
Tier-0 and Tier-1 Supercomputers
Reproducible software deployment using Guix on French Tier-1 and EuroHPC Tier-0 machines is one of the goals of the Development and Integration project of NumPEx (ExaDI), the French exascale HPC program.
As highlighted in last year’s report, Guix being unavailable so far on
these supercomputers, our focus has been ensuring that the HPC software
stack packaged in Guix, bundled as a tarball or Apptainer image produced
by guix pack, runs flawlessly on these machines. This is now
well supported for all the French Tier-1 clusters and for several
EuroHPC supercomputers (LUMI, MeluXina, Vega, etc.), including for the
variety of high-speed interconnects and GPUs found on these machines.
We reported on this work in a blog
post
and gave a
webinar
and a conference
tutorial.
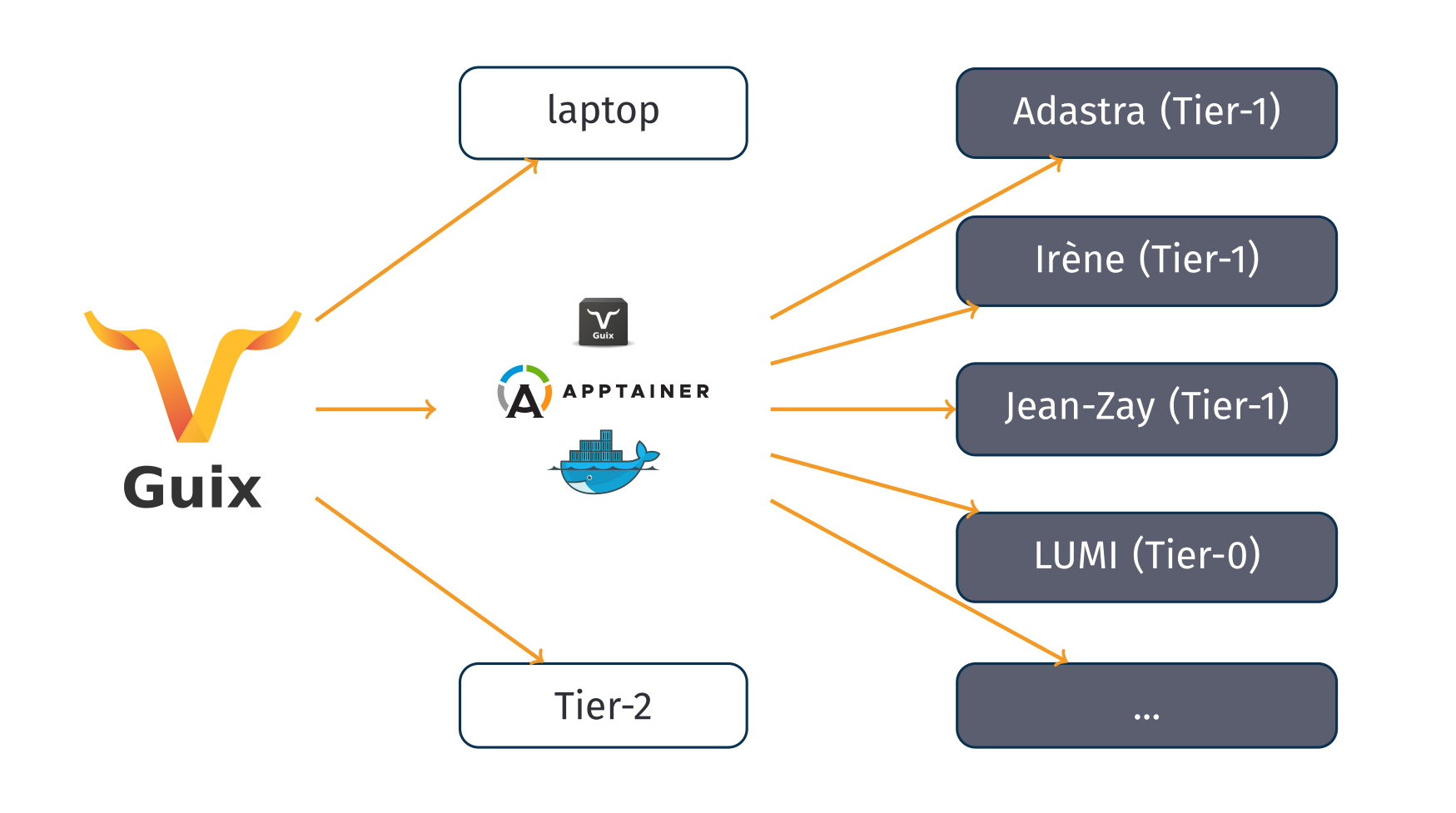
Overall this leads to a situation where, so far, one may use Guix
directly on their laptop (for development purposes) and on a variety of
Tier-2 clusters, resorting to the tarballs and Apptainer images
produced by guix pack when targeting supercomputers where Guix is
currently missing.
In early 2025, ExaDI held a meeting with the HPC department of CINES, the French Computing center for higher education and research, operator of Adastra, one of the three Tier-1 supercomputers in France. CINES wants to provide users with Guix as an alternative software deployment method giving access to a large software catalog and work is underway to make it a reality. Guix is expected to be made available to Adastra users in the first quarter of 2026.
Running the Build Daemon Without Root Privileges
Work with CINES triggered new developments in Guix, allowing the build
daemon, guix-daemon, to run without root privileges. Traditionally,
guix-daemon would run as root and use these privileges to spawn build
processes in isolated (and unprivileged!) build environments—thereby
maximizing the changes that builds are fully reproducible. Having a
daemon running as root, even isolated in a separate node or virtual
machine, is often met with reluctance on the part of cluster system
administrators.
The rootless mode builds on unprivileged user namespaces and
associated Linux process isolation mechanisms that, until recently, were
disabled on many supercomputers. Development required careful review:
the isolation mechanisms used when guix-daemon runs as an unprivileged
user are very different from the more traditional mechanisms used when
it runs as root. Furthermore, early experimentation revealed in some
cases discrepancies between the build environment set up by the rootless
daemon compared to the root daemon. We believe those have now been
addressed.
We covered the rationale and development of the rootless build daemon in a blog post earlier this year. The rootless mode addresses what many administrators may have perceived as a blocker.
Pangenomics and Genetics Research Cluster at UTHSC
At the University of Tennessee Health Science Center (UTHSC), Memphis TN (USA), we are running a 16-node large-memory
Octopus HPC cluster (438 real
CPU cores) dedicated to pangenome and genetics research. In 2025, we replaced the LizardFS distributed network storage with a modern MooseFS. MooseFS already proves a great improvement because it allows for multiple data storage strategies using a single mounted directory on each node. For example the /moosefs/scratch directory allows for multiple redundant copies of files that a distributed striped 'torrent' fashion across the cluster and files in /moosefs/tmp exist without any redundancy at all. The no-redundancy option is not available in Ceph and (obviously) allows for more data on the cluster. That is why we decided to go for MooseFS. Currently, we are implementing network boot for the cluster which will allow us to run recent Debian nodes, addressing the 'old distro' upgrade issue that we often encounter in HPC environment. It also opens the way to introducing pure Guix nodes or VMs. Finally, we are upgrading the optical network to increase speeds on our HPC.
Notable about this HPC cluster is that it is administered by the users themselves. Thanks to Guix, we install, run and manage the cluster as researchers — and roll back in case of a mistake. All critical services, such as MooseFS and Slurm are running as versioned Guix deployments. In addition to keeping our sanity that even allows for (temporary) using different versions of distros on the nodes. UTHSC IT manages the infrastructure—i.e., physical placement, electricity, routers and firewalls — but beyond that there are no demands on IT. Thanks to out-of-band access, we can completely (re)install machines remotely. Octopus runs Guix on top of a minimal Debian install and we are experimenting with pure Guix virtual machines and nodes that can be run on demand. Almost all deployed software has been packaged in Guix and can be installed on the head-node by regular users on the cluster without root access. This same software is shared through NFS on the nodes. See the guix-bioinformatics channel for all deployment configuration.
Thanks to Collin Doering and the wider Guix community since 2024 we now have Guix on a bare metal machine that acts as a public build 'farm' for Guix North America that serves the continent. Collin's blog goes into more detail in his post Setup of a Simple Guix Build Farm and Substitute Server.
RISC-V Progress
Christopher Batten (Cornell) and Michael Taylor (University of Washington) are in charge of creating the NSF-funded RISC-V supercomputer with 2,000 cores per node and 16 nodes in a rack (NSF PPoSS grant 2118709), targeting Guix driven pangenomic workloads by Erik Garrison, Arun Isaac, Andrea Guarracino, and Pjotr Prins. We have a prototype running!
The supercomputer will incorporate Guix and the GNU Mes bootstrap, with input from Arun Isaac, Efraim Flashner and others.
NLNet funds RISC-V support for the Guix riscv64 target from Efraim Flashner and the GNU Mes RISC-V bootstrap project with Ekaitz Zarraga, Andrius Štikonas, and Jan Nieuwenhuizen. The RISC-V bootstrap is fully working and can be adopted by Linux distributions! People started running GNU Guix on RISC-V bare metal.
Outreach and User Support
Guix-HPC is in part about “spreading the word” about our approach to reproducible software environments and how it can help further the goals of reproducible research and high-performance computing development. This section summarizes talks, training sessions, and related activities this year.
A Busy Year Upstream
Upstream, in the Guix project itself, it’s been a busy year. In early 2025, the community started discussions to migrate its development platform. This led to the decision to migrate, recorded in a Guix Consensus Document (GCD). The plan described in this GCD was implemented in May 2025, with Codeberg becoming Guix’s development platform. Guix-Science made the same move earlier, in January 2025.
Building on the experience of Guix-Science, Guix has set up an instance of the Cuirass continuous integration (CI) tool that automatically builds pull request branches. This is still a work in progress but it has already proved to be helpful.
Since January 2025, almost 450 people contributed to Guix itself—a 20% increase compared to the previous year. The switch to a pull-request workflow with continuous integration may have been viewed by many as an improved contributor experience.
User and Contributor Survey
As the Guix project continues to grow and evolve, with new contributors and users joining, a Guix User and Contributor Survey was conducted in early 2025, spearheaded by contributor Steve George, to better understand the perspectives and experiences of the community.
This survey served as an effective tool for gathering high-quality feedback from a diverse segment of the community. Although many topics could have been explored, the focus was specifically on understanding how Guix is used, how contributors interact with the project, and what their expectations are. The results, along with related anonymised data, were released under the Creative Commons CC0 licence, enabling anyone to conduct further analysis and derive additional insights.
Almost one thousand participants with a wide range of experience levels took part in the survey. Most were recent users (within the last five years), primarily motivated by Guix's declarative configuration and reproducibility features, and reported a largely positive adoption experience. Notably, the majority of users rely on Intel/AMD hardware, with a significant number also using Arm (AArch64) architectures.
It is evident that Guix thrives within a dynamic community, where most users are also active contributors, engaging in development through submitting patches, reviewing code, and reporting bugs. The new Codeberg organization for code development and issue tracking is expected to further enhance this collaborative spirit.
Articles
The following refereed articles about Guix were published:
- Lars Bilke, Thomas Fischer, Dmitri Naumov, and Tobias Meisel, Reproducible HPC software deployments, simulations, and workflows — a case study for far-field deep geological repository assessment, Environmental Earth Sciences, August 2025
- Dylan Bissuel, Léo Orveillon, Benjamin Arrondeau, Paulo Almeida De Mendonça, Irina Piazza, Martin Uhrin, Étienne Polack, Akshay Krishna Ammothum Kandy, David Martin-Calle, Jonathan Chapignac, Aadhityan Arivazhagan, Lorenzo Paulatto, Pierre-Antoine Bouttier, M.-I Richard, Thierry Deutsch, David Rodney, A M Saitta, Nöel Jakse, Reproducible container solutions for codes and workflows in materials science, December 2025
- Simon Tournier, Guix: From Transparent and Verifiable to Long-Term Reproducible Research?, DebConf25 Brest, IRISA, July 2025
Talks
Since last year, we gave the following talks at the following venues:
- Guix + Software Heritage: Source Code Archiving to the Rescue of Reproducible Deployment, Open Research Tools and Technology track, FOSDEM, Feb. 2025 (Simon Tournier)
- Guix: toward practical transparent, verifiable and long-term reproducible computational environment, seminar IBPC, Mar. 2025 (Simon Tournier)
- Traçabilité des environnements logiciels avec Guix, internal seminar of MOABI / Seqoia team of AP-HP, Jun. 2025 (Simon Tournier)
- Guix: From transparent and verifiable to long-term reproducible research? DebConf, Jul. 2025 (Simon Tournier)
- Hands-on reproducible computing with Guix (video), RSECon, Sept. 2025 (Ghislain Vaillant, Julien Castelneau)
- What about reproducible computational environments? (video), workshop RT Santé Numérique, Oct. 2025 (Simon Tournier)
- Guix for FPGAs, webcast, OrConf 2025, Sep. 2025 (Cayetano Santos)
Events
Guix and Nix miniconf, Nov. 2025, Toulouse (satellite of Capitole du Libre)
As has become tradition, Pjotr Prins and Manolis Ragkousis spearheaded the organization of the “Declarative and minimalistic computing” track at FOSDEM 2025, which was home to several Guix talks, along with the satellite Guix Days where 50 Guix contributors gathered.
Training Sessions
Café Guix
Café Guix is an informal monthly discussion about the Guix software environment manager. Students, researchers, system administrators, support staff from laboratories or computing centres — everyone is welcome to join in this one-hour monthly online meeting to discuss questions they have about Guix and its use in the broadest sense. Each session has a specific level of difficulty, clearly indicated in the program. Some sessions are designed for beginners and others for more experienced users. For example, in 2025, entry-level topics included an introduction of Guix from a user point of view and the presentation of basic commands. The question of short- and medium-term reproducibility was also discussed. We also covered more advanced topics such as the use of guix pack, to efficiently reproduce your software environment on a machine that does not have Guix installed. Each session is recorded and slides and video are available from the web page.
Compas conference
As part of Compas, the annual conference of the French HPC, system and architecture scientific communities, we organized two tutorials: the first one, led by Marek Felšöci, showed how to get started with Guix and to combine in with Org-Mode for literate programming, and the second one, led by a team at Inria Bordeaux, showed how to take advantage of Guix to run code on national supercomputers. Material for both tutorials is available online.
MOOC “Reproducible Research II: Practices and tools for managing computations and data”
A second session of this online course (MOOC) has been run from 5 May to 30 September 2025. One of its three modules is about reproducible computational environments. It includes a detailed introduction to Guix, with exercises, and it uses Guix to ensure the reproducibility of its main example, a workflow for detecting sunspots in a database of thousands of images of the sun. The Guix material has been thoroughly revised for this second session, in particular by reworking the exercises for improved clarity. A revised and extended third session will open on 5 May 2026 in self-paced mode, meaning that it will be open for a long period (at least until the end of 2026), with students being able to join at any time and advance at their own pace.
NumPEx webinar
One of the objectives of the Development and Integration work package of NumPEx, the French exascale HPC program, is to provide training for the French HPC community. A webinar titled "Introduction to Guix" (supporting material) has been presented and recorded (to be moved to Canal-U.tv).
RSECon hands-on
This workshop provided a guided tour of Guix, focused on introducing the key concepts and commands of this tool, and getting hands-on experience with its practice. First, people has been introduced to the Guix package management commands, created a reproducible environment with
guix shell, and deployed it withguix time-machine. Then, the session walked through the process of “Guixifying” a representative scientific pipeline from the neuroimaging community. The session ended with a broad discussion on Guix adoption in HPC, some of its advanced features, and available support channels.All the material is available online.
SantéNum hackathon
The mission of Digital Health Priority Research Program (PEPR Santé Numérique) is to federate a national, multidisciplinary research community active in digital health. In this context, interdisciplinary research groups are creating new or innovative approaches for processing health data. They require reproducible computational environments. The 2.5 days of hands-on had been an opportunity to dive into core concepts of Guix and to package dedicated software involved in Digital Health.
Preparing and running the session led to the package inclusions by seasoned contributors in the Guix-Science third-party channel. The outcome of the training session is contributions by newcomers.
Personnel
GNU Guix is a collaborative effort, receiving contributions from more than 90 people every month. As part of Guix-HPC, participating institutions have dedicated work hours to the project, which we summarize here.
- CNRS: 0.2 person-year (Konrad Hinsen)
- Inria: 4 person-years (Ludovic Courtès and Romain Garbage; contributors to the Guix-HPC, Guix-Science, and Guix channels: Emmanuel Agullo, Julien Castelneau, Luca Cirrottola, Gilles Marait, Florent Pruvost, Philippe Swartvagher, Ghislain Vaillant; system administrators in charge of Guix on the PlaFRIM and Abaca/Grid’5000 clusters: Julien Lelaurain, Philippe Virouleau)
- University of Tennessee Health Science Center (UTHSC): 3+ person-years (Efraim Flashner, Collin Doering, Bonface Munyoki, Fred Muriithi, Arun Isaac, Andrea Guarracino, Erik Garrison and Pjotr Prins)
- GRICAD : CNRS 1 person-year (Benjamin Arrondeau, Pierre-Antoine Bouttier, Loris Mégy and Léo Orveillon), UGA 0.1 person-year (Céline Acary-Robert)
- Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC): 1 person-year (Ricardo Wurmus and Mădălin Ionel Patrașcu)
- Université Paris Cité: 0.5 person-year (Simon Tournier)
Perspectives
Scientists from a growing number of domains are adopting Guix, as illustrated by the introduction of new packages in the Guix-Science channel. It is difficult but necessary to encourage package creators to validate their Guix packages upstream in order to increase the size of the Guix community and promote multidisciplinary exchanges. Perhaps now is the time to better coordinate and structure the development of packages in order to improve the pooling of efforts.
Guix-HPC will continue its support and training activities on reproducibility issues, by organizing awareness-raising events and workshops—such as the upcoming session in Bordeaux in 2026. Sharing experiences about Guix in various scientific domains with a focus on the central theme of software environment reproducibility encourages people to improve the way they work and run their computational experiments.
We anticipate that promoting and spreading the use of the guix pack functionality together with the possibility of running guix-daemon as an unprivileged user will encourage the use of Guix in more national and regional clusters. Moreover, participation in the WIDERA program of the Horizon Europe project will foster European collaboration to promote its use.
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).





