Towards reproducible Jupyter notebooks
Jupyter Notebooks are becoming a key component of the researcher’s toolbox when it comes to sharing and reproducing computational experiments. Jupyter notebooks allow users to not only intermingle a narrative with supporting code in a way reminiscent of literate programming, they also make it easy to interact with the code and, thus, build on the work of each other.
Update: My FOSDEM 2020 talk, as well as the slides and video of my talk (in French) at JCAD 2019 are available on-line.
To give a few examples, Jupyter notebooks are one of the topics of Inria Learning Lab’s new MOOC on reproducible science and a central part of the “Reproducible Science Curriculum”; researchers in different domains view Jupyter notebooks as the tool of choice to support reproducible research and, possibly, as the seed for changes in scientific publication practices.
In this post we explore a solution to Jupyter Notebook’s Achille’s heel: software deployment. We have been working on a solution to this dubbed Guix-Jupyter that we’re releasing today, but first, let us explain how Jupyter’s promises for reproducible research are hindered by a lack of support for reproducible software deployment.
The extent of Notebook reproducibility
A Jupyter Notebook is essentially a program, and the jupyter notebook
command provides an interactive user interface in your web browser to
that program. Of course, to execute the program, an interpreter and a
Jupyter
kernel
for the language the program is written in must be available in the
execution environment of Jupyter. If the program refers to external
libraries or data, well, those also have to be available in the
execution environment.
Unfortunately, it is up to you, the user, to run Jupyter in the right environment, with the right interpreter and libraries available. Say I publish a notebook containing a Python 3 program that uses NumPy and SciPy. If you try to run it on a Jupyter instance that only has Python 2, or that lacks SciPy, it won’t run; if you have SciPy but a different version that the one I used, well, it may fail to run because of API changes, or you could get different results.
What’s worse, Jupyter does not offer a standard way for notebooks to express their software dependencies. You can only hope the author provides detailed instructions and then walk your way through deployment.
Simple solutions!
People have got used to resorting to a simple solution: running pip install or conda install right at the top of their
notebook!
This isn’t great for several reasons. As a researcher, you’re
interested in reproducing or deriving someone else’s research; you’re
certainly much less interested in installing arbitrary packages on your
system as you do that. Plus, that doesn’t necessarily quite solve the
dependency issue: pip can only install Python dependencies, not Python
itself or anything written in another language; the JupyterLab Package
Installer
streamlines the pip install trick but doesn’t address this core
limitation. conda doesn’t have this drawback, but it still assumes
the availability of “system software” packages, and is generally not
very good at reproducing software environments at different points in
time or
space.
I hear you… we have our next go-to solution: containers to the rescue! That is true, thanks for suggesting it! Indeed, many people have developed solutions to the Jupyter deployment problem around containers. As a user, you can entrust a service such as Binder with your code and data, which conveniently takes care of building a Docker image for the software environment of your notebook (using the nifty repo2docker) and spawning a Jupyter Notebook instance in that environment.
Research institutes and computing centers have also started offering “Jupyter Notebook as a service” in a similar way. Administrators of those systems can “just” deploy JupyterHub with Kubernetes on OpenStack, possibly with a drop of BinderHub for good measure.
Mind you, this software stack achieves an amazing job, but its complexity is baffling if we think about the simple deployment problem we’re trying to solve. And maybe you don’t want to turn your computers into mere Web terminals when you could run notebooks locally.
Last but not least, we still haven’t solved the core issue, which is
that notebooks are not self-contained: they do not describe the
dependencies they need. Binder’s configuration
files,
such as environment.yml for Anaconda, get us close to that, but they
fail to capture a complete environment, thereby making it hard to
impossible to reproduce the same environment on different machines or at
different points in time.
Making Notebooks “deployment-aware”
What if we could make notebooks “deployment-aware” from the start? What if the notebook itself could describe its dependencies? What if reproducible software deployment was an integral part of the notebook?
We started working in that direction a year ago when Pierre-Antoine Rouby wrote a first version of the Guix kernel for Jupyter.
Today, we’re happy to announce the first beta release of the Guix-Jupyter, a Guix kernel for Jupyter!

The Guix kernel is still very much a work-in-progress but it already lays the foundation for self-contained, reproducible notebooks—notebooks that automatically run in the right software environment, regardless of the machine where you run it or the time at which you run it. We’re pretty excited to share it today!
So, what does the Guix kernel have to offer? First and foremost, it
allows you to define environments in which the notebook code is going
to be executed. An environment consists of any number of Guix
packages and one of them must be a
Jupyter kernel—e.g.,
python-ipykernel for
Python 3 or r-irkernel for
GNU R. And of course, you can add any Python or R libraries or really
any package you need to use in those environments. Subsequent cells are
automatically executed in that environment, using the Jupyter kernel it
contains.
In fact, a single notebook can define several environments, each with a possibly different Jupyter kernel, which allows you to create a multi-lingual notebook:
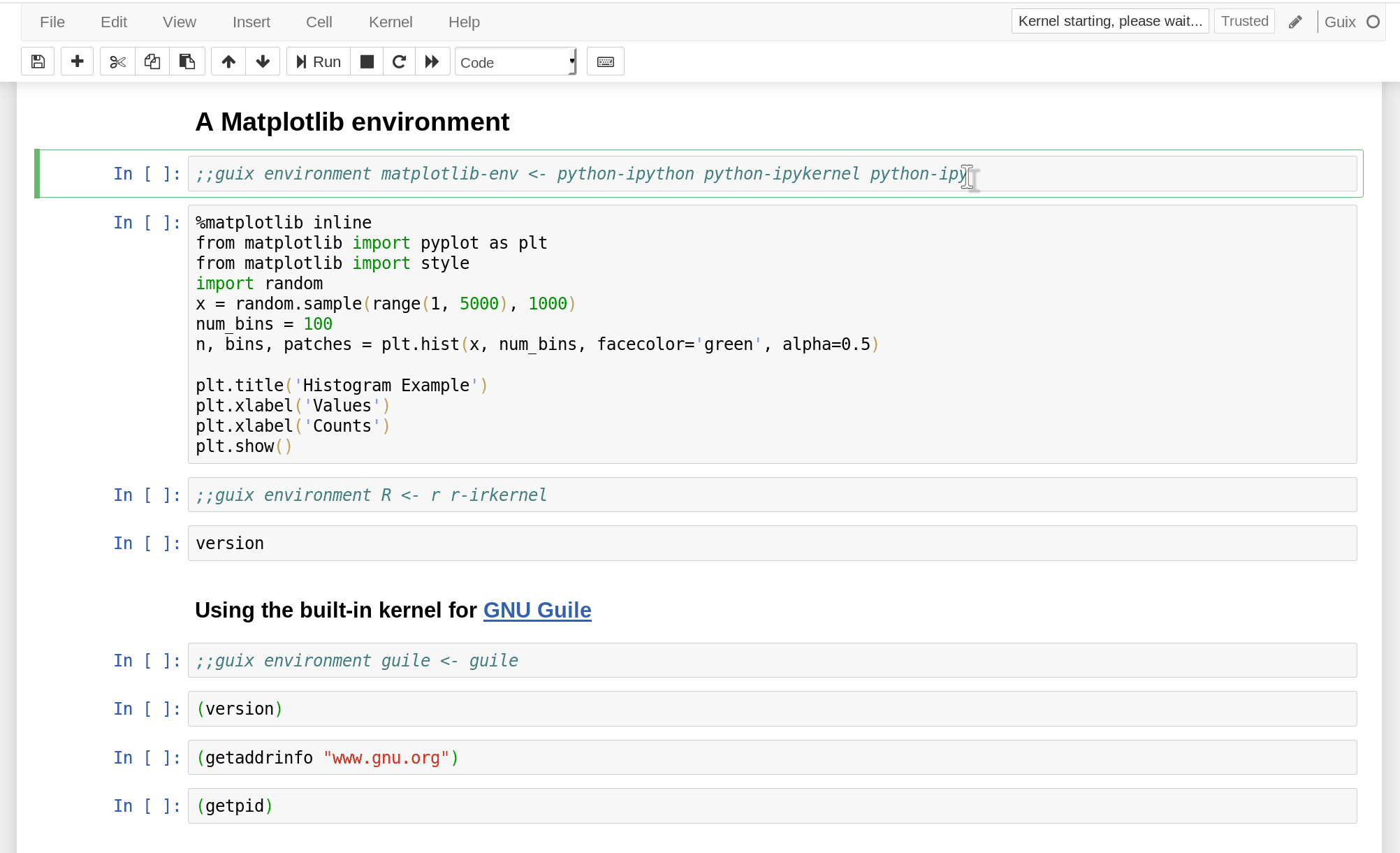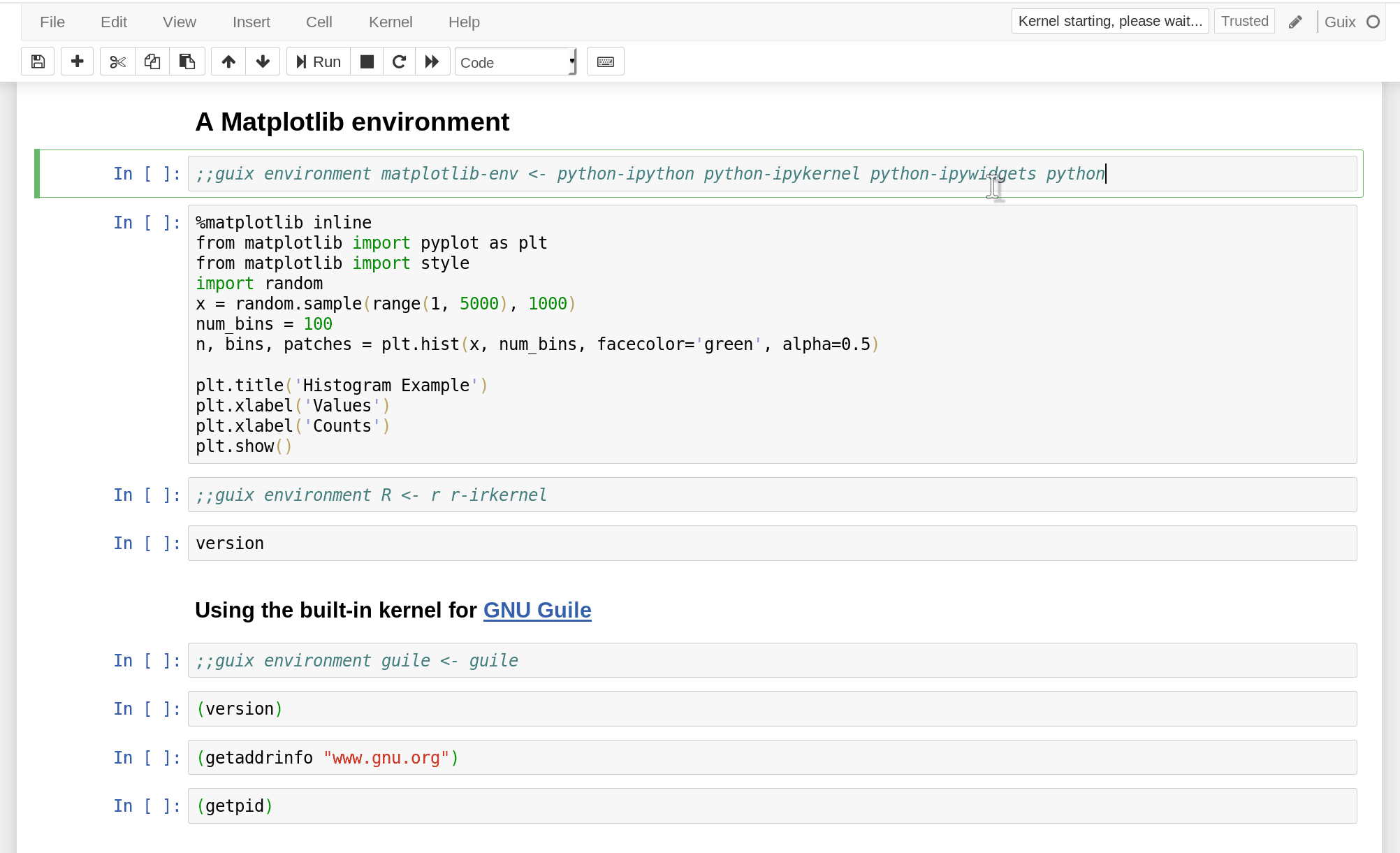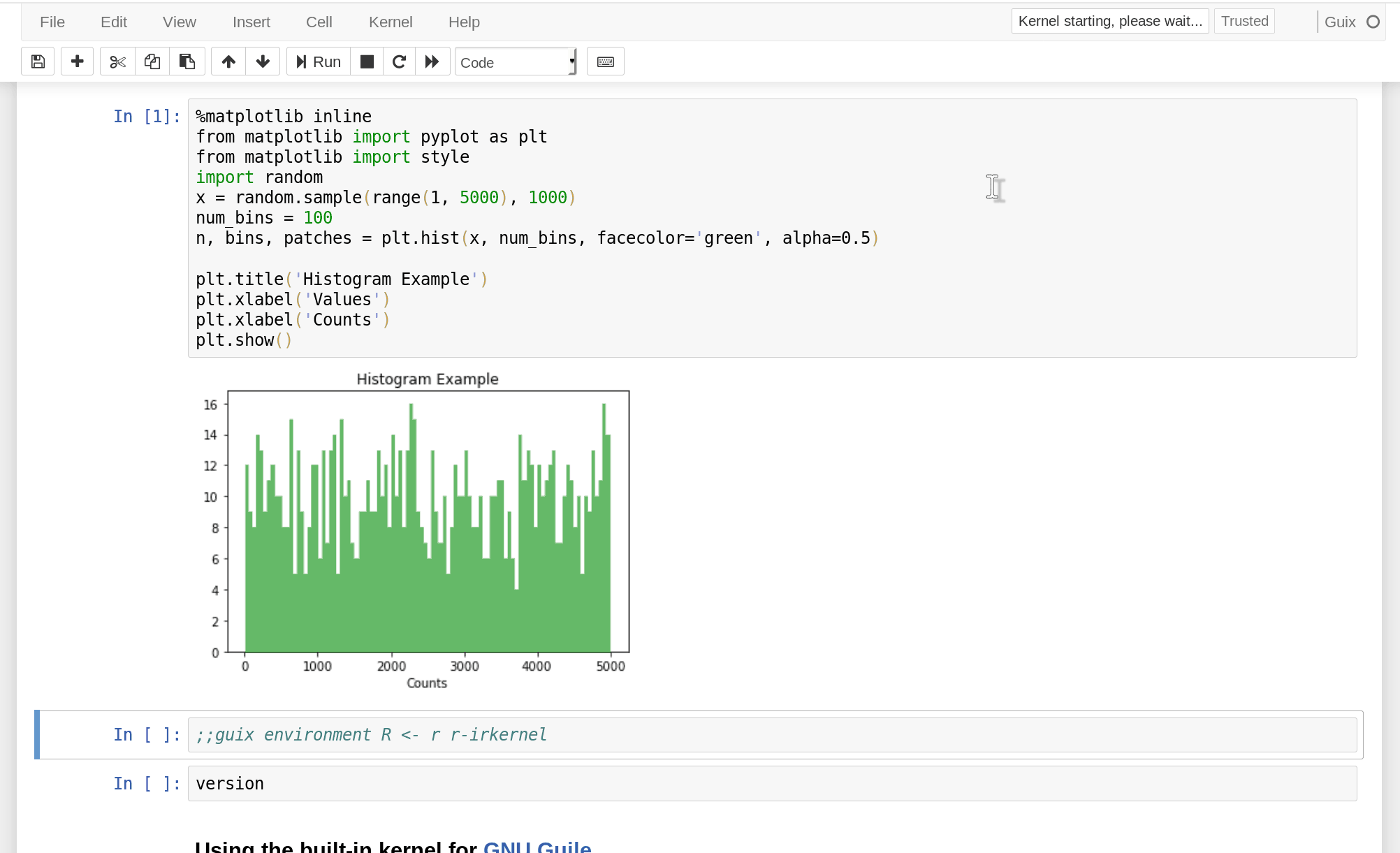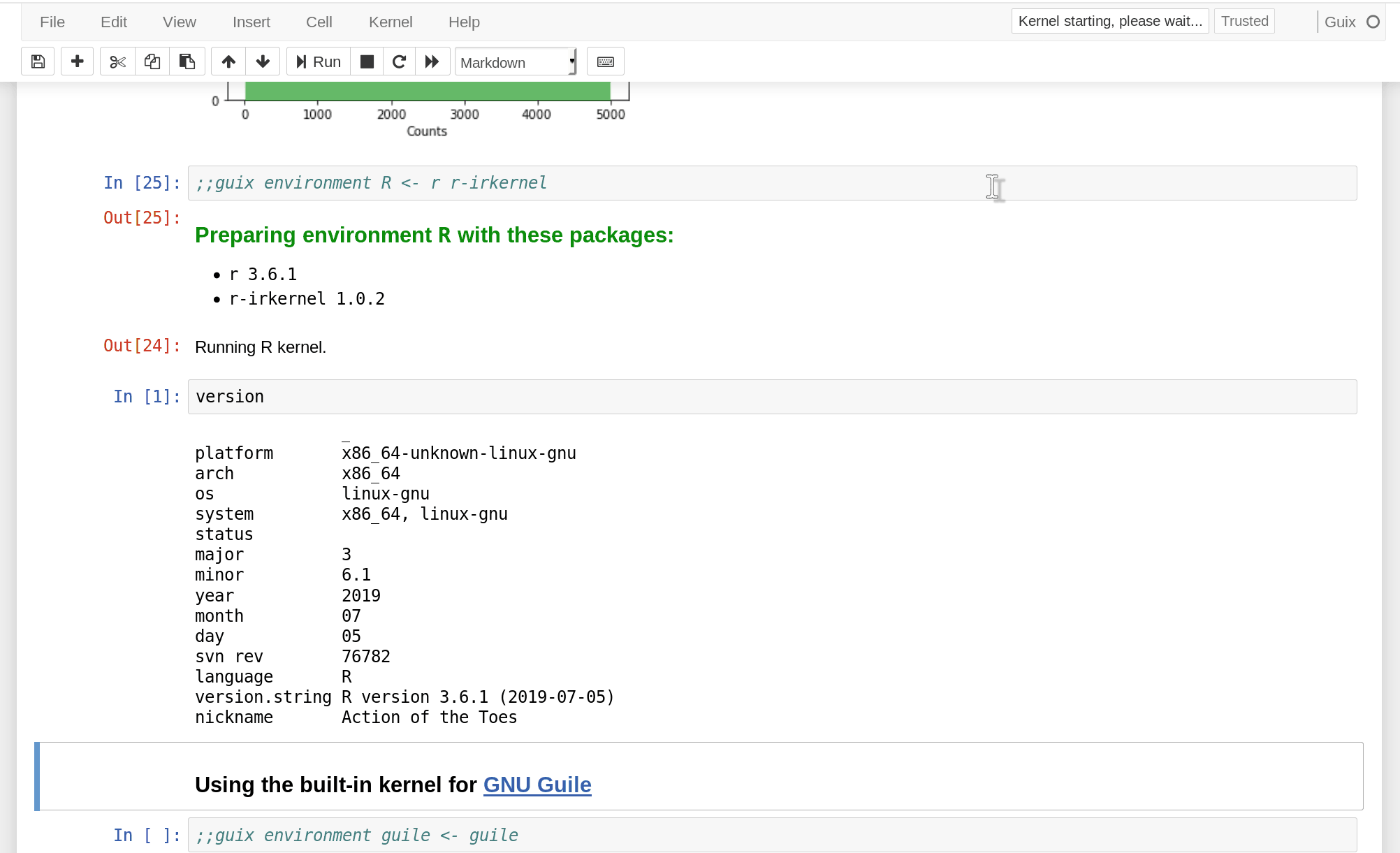
(The IPython kernel has a built-in mechanism to interface with languages other than Python, but that’s a wholly different approach.)
How does that differ from running pip install or similar right from
the notebook? First, it doesn’t fiddle with your home directory or
similar—the environments are one-off environments created on the fly.
Second, it’s not limited to a particular language. And third, it’s
reproducible.
Namely, since Guix is able to reproduce software environments at any point in time and space, you can not only specify packages to include in the environment, but also pin a specific revision of the Guix channels:
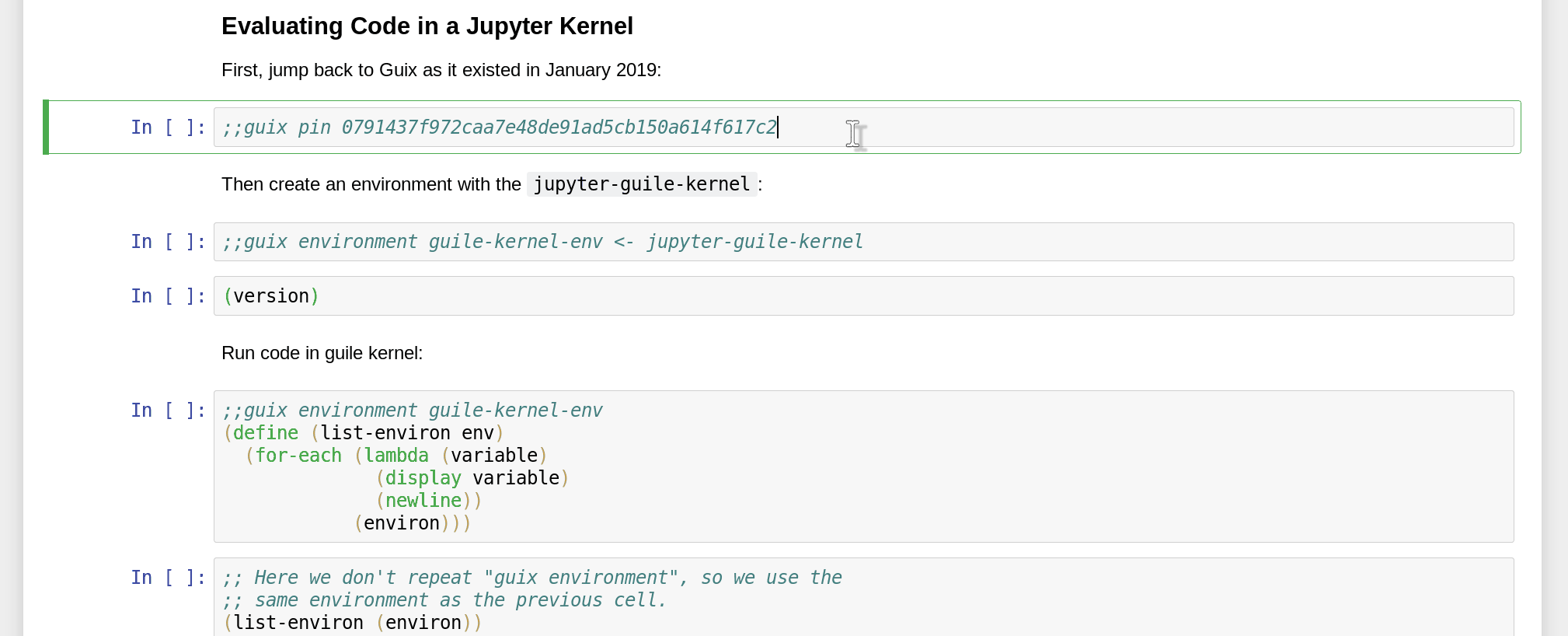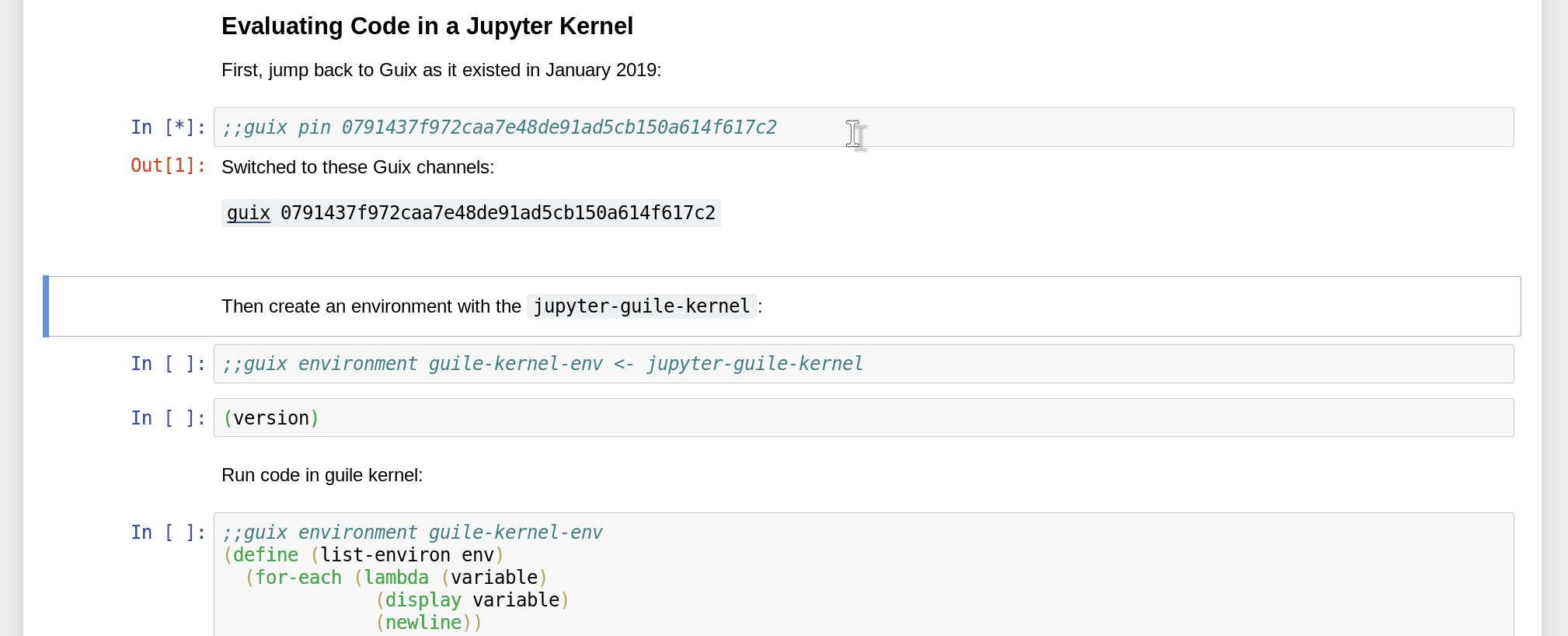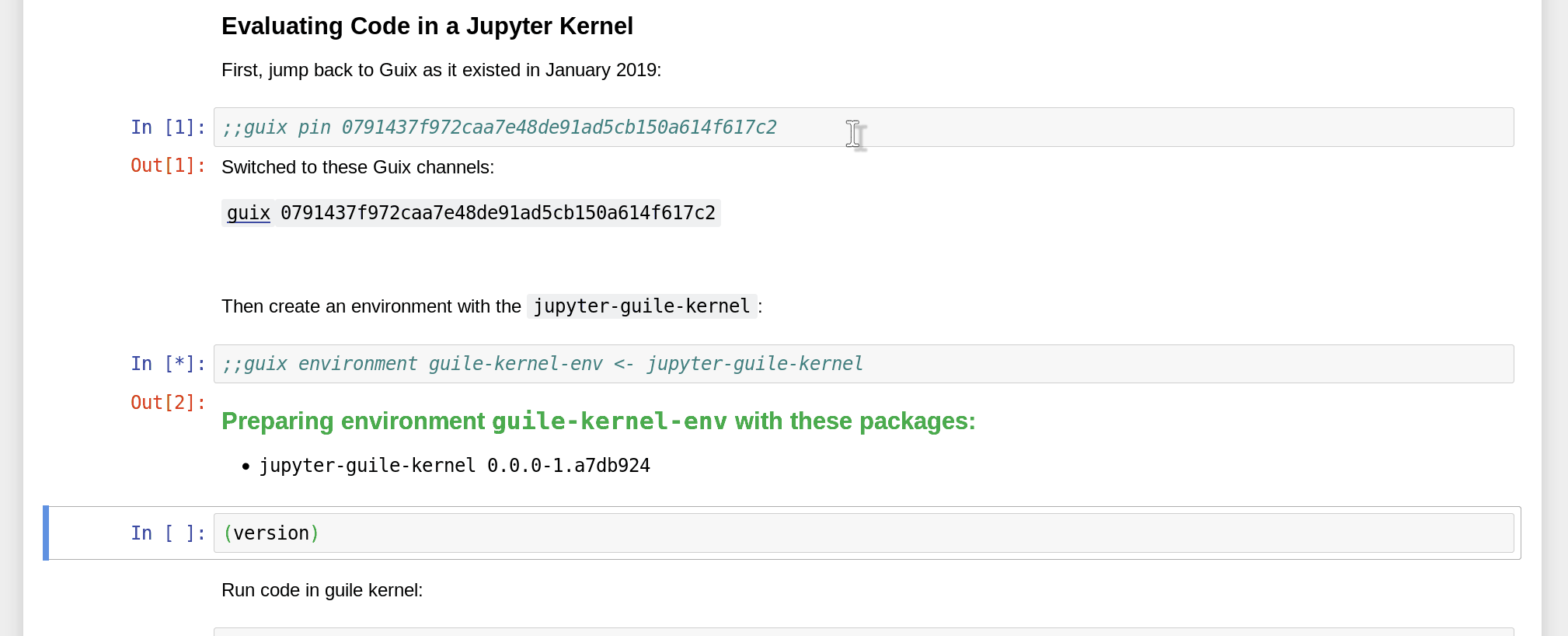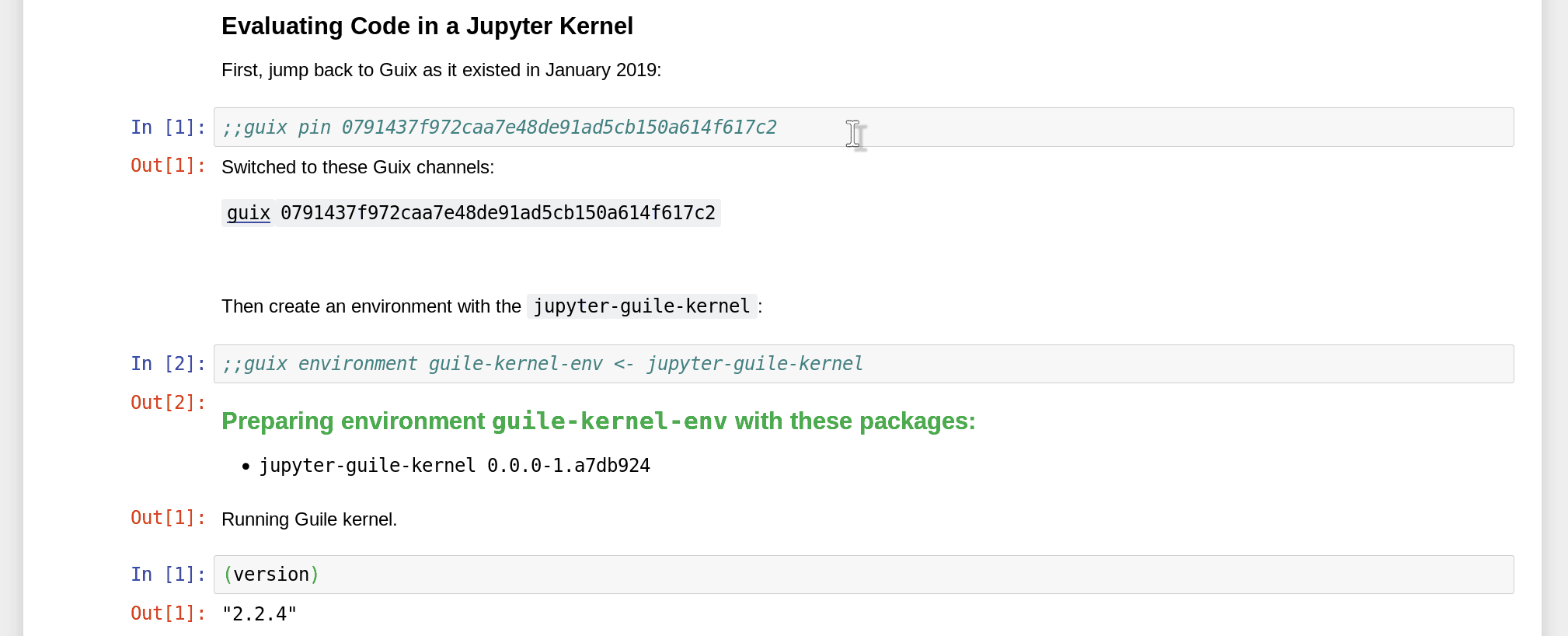
How do you obtain the commit ID that you want to pin to in the first
place? If you’re using Guix, you can obtain it by running guix describe
on a configuration that works for you. Beyond that, the brand-new Guix
Data Service will come in handy. For
example, it can show you the history of upgrades to any given package,
say
python-scipy.
By hooking up Guix-Jupyter and the Data Service, we could make it easier
to do time traveling. We’ll see!
Isolated execution environments
What’s more, the notebook code runs in an isolated environment: it cannot access any of your files and cannot fetch data from the Internet (more on the implementation below). That’s good for security (you can now run untrusted notebooks locally), but that’s also good for reproducibility: the notebook cannot have undeclared dependencies. In fact, we’re adapting the functional model of build processes pioneered by Nix to an interactive execution environment. In other words, we’re saying that a reproducible notebook is a pure function, and we create an isolated execution environment to make it happen.
So far so good, but if you’ve payed attention, you’re probably
wondering: how do I get my data in that environment? To refer to data,
the notebook must use a ;;guix download directive containing a URL and
expected SHA256 hash of the data:
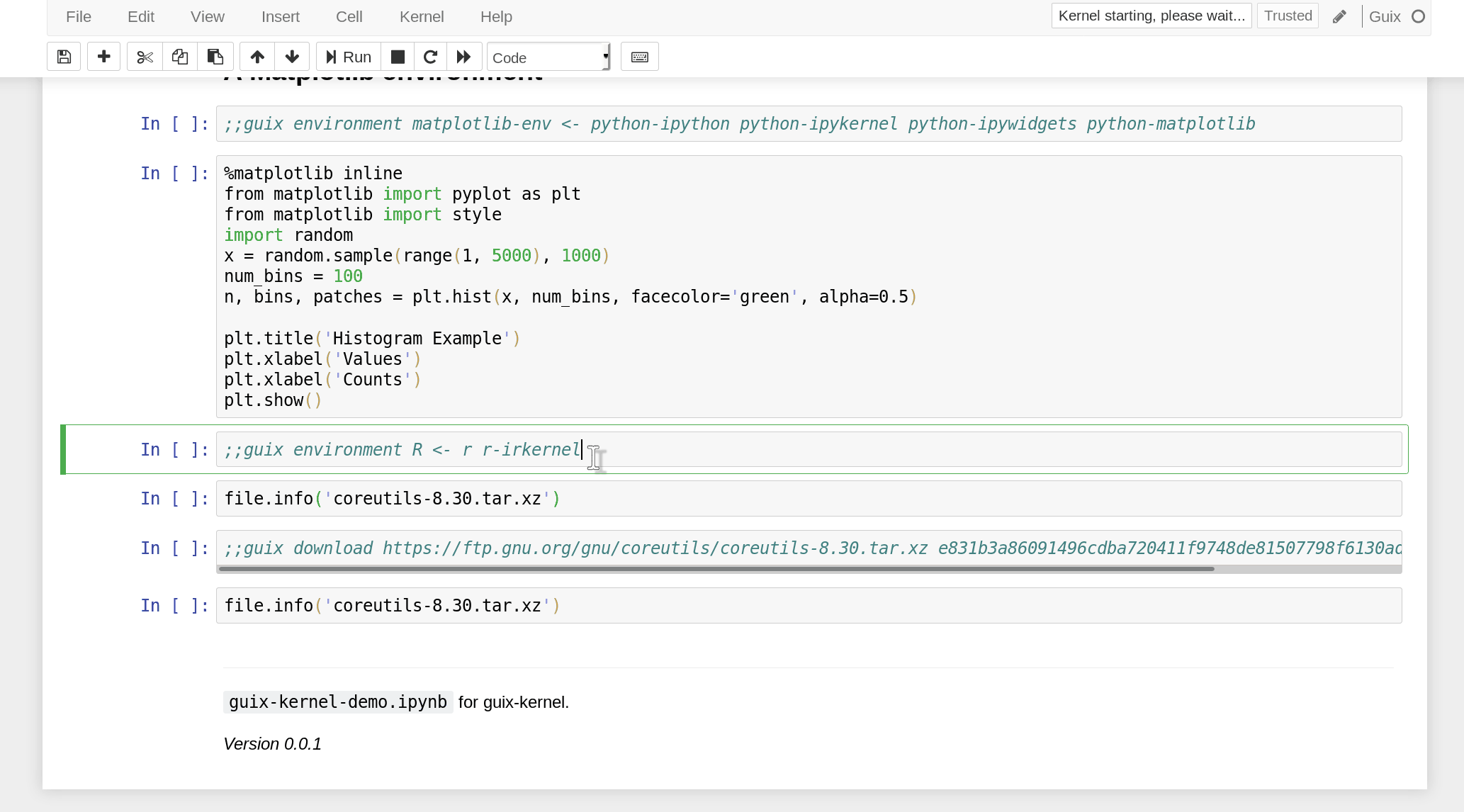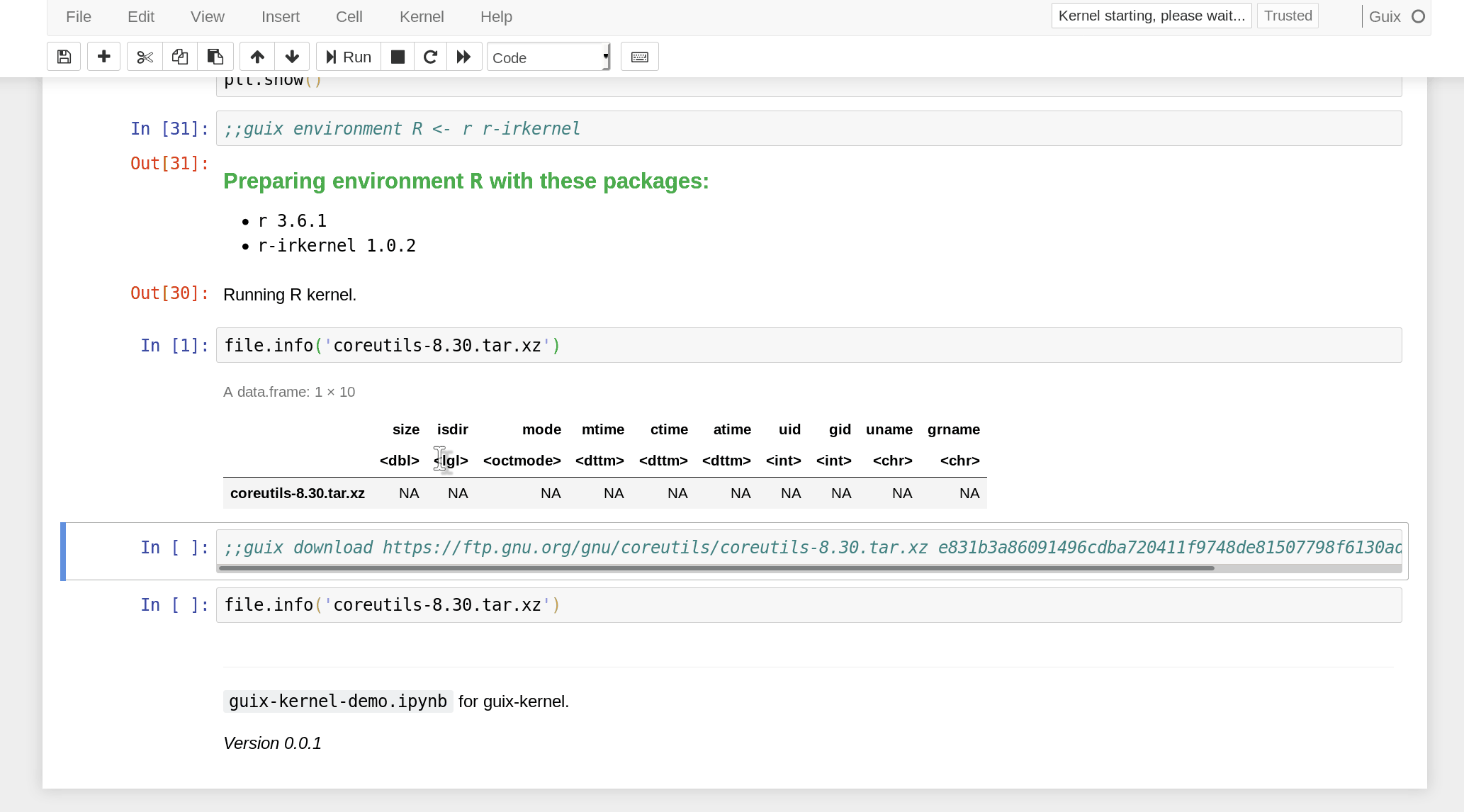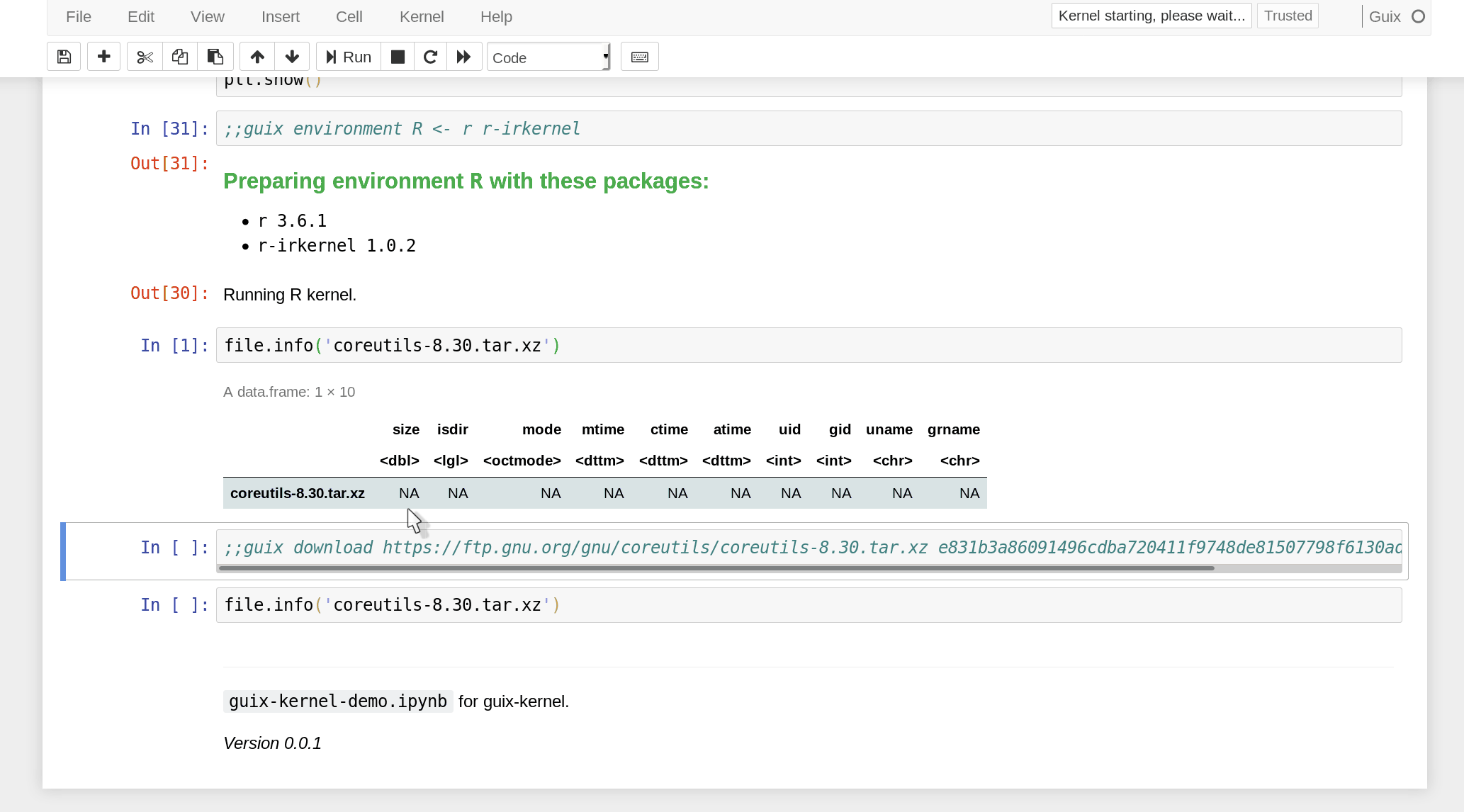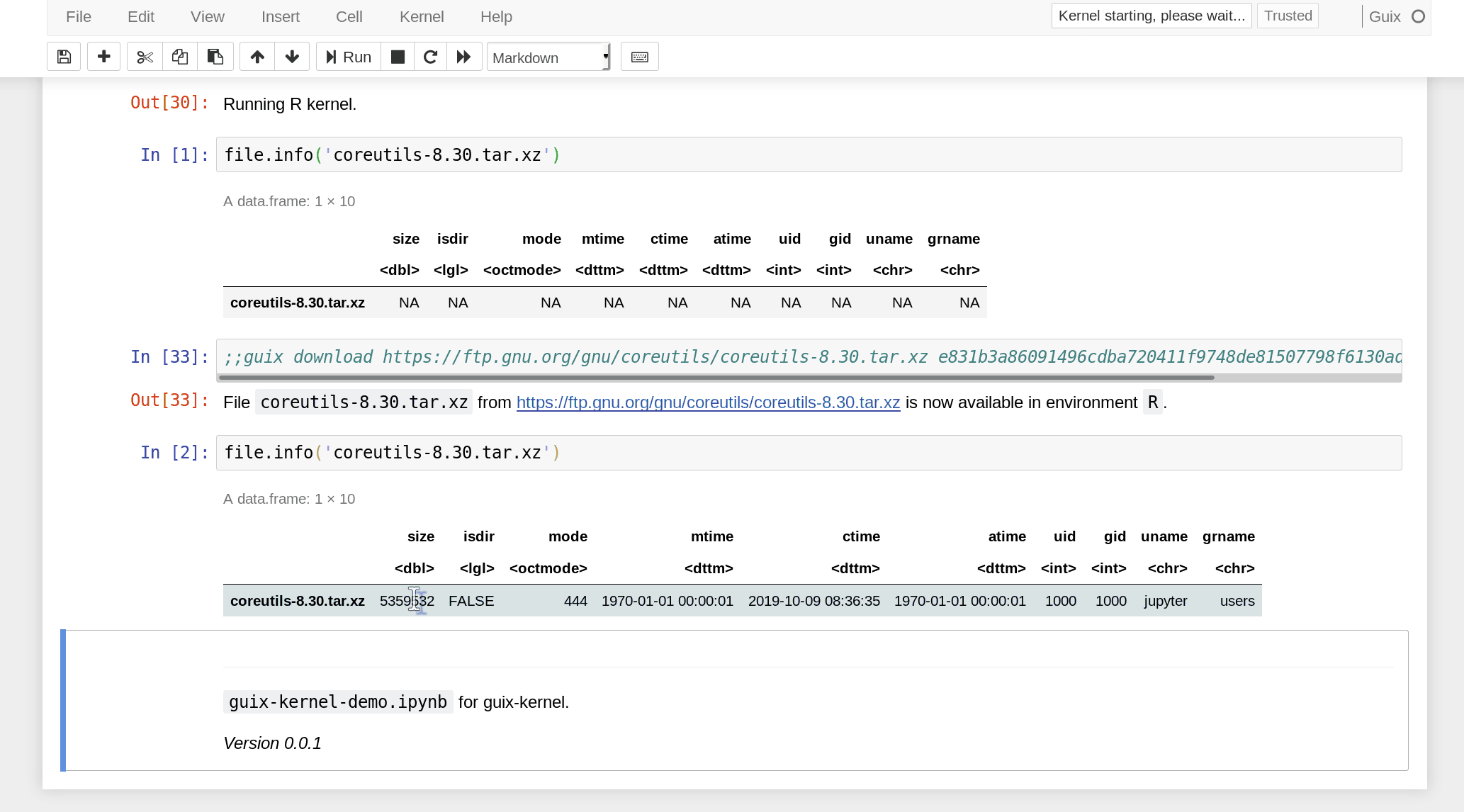
In practice, data is only downloaded the first time. Subsequent executions reuse the pre-downloaded data. In Nix/Guix terms, this is a “fixed-output derivation”. Since the hash of the data is specified, we make sure the notebook operates on the intended data, and an error is raised if the downloaded data has a different hash.
If you’ve used Guix before, the ;;guix annotations are similar to the
interface of command-line tools like guix environment,
guix pull,
and guix describe.
Guix kernel architecture
The Guix support for Jupyter we presented above is not implemented as a standard Jupyter extension, but rather as a Jupyter Kernel that stands alone and acts as a proxy between the clients and the actual kernels you use.
Thus, as a user, you first have to install Guix on your machine, and then Jupyter and the Guix kernel:
guix install jupyter guix-jupyterAt that point, you can start a notebook:
jupyter notebook… and select the “Guix” kernel.
Then you don’t need to explicitly install any other Jupyter kernel since
you can just add them to your notebook via ;;guix environment
annotations. That’s the nice thing about implementing it as a kernel.
Technically, the kernel implements all the Jupyter messaging
protocol
in Guile Scheme, in a type-safe way: JSON messages are converted to
Scheme records and
back,
which allows us to catch certain mistakes at compile time. There kernel
maintains
state,
such as the list of environments and proxied kernels running. It
inspects execute_request messages to see if they might contain a
;;guix magic, handles that if needed, and otherwise passes them on to
the relevant proxied kernel. Other messages such as complete_request
(for code completion) are treated similarly. Processes in separate
namespaces
are created using Guix’s container
API.
As a bonus, there’s, of course, a built-in kernel for GNU Guile, the great Scheme implementation that powers Guix. Pictures, relational programming, delimited continuations, and whatnot in your notebook!
One downside to the proxying approach is that since a notebook is normally monolingual, there’s no way to tell Jupyter that some cells are Python, while others are R, Guile, and so on.
It must be said though that we’re much more familiar with Guix than with Jupyter. So if you’re a Jupyter hacker, do share any piece of advice you may have!
Conclusion
The Guix Jupyter kernel is still “beta” but it already demonstrates most of the things we had in mind when we toyed with the idea of “notebooks with reproducible deployment built-in”. There’s many improvements we can make, notably to the user interface: things like showing a progress bar when an environment is being built, providing widgets to navigate environments or packages, etc.
It remains to be seen how convenient Guix-Jupyter is for “real-world”
notebooks, and we’d very much like to hear from intrepid Jupyter users
who’d want to try and add ;;guix annotations to their favorite
notebooks.
A practical question is: what happens if you publish a notebook for the
Guix-Jupyter kernel but your collaborators don’t have that kernel? If
your notebook uses a single environment (say, a single Python
environment), they’ll be able to run it provided they remove or skip the
;;guix annotations. But then, of course, they’re on their own when it
comes to deploying the environment of that notebook. If you use ;;guix download or multiple environments, then the notebook won’t be readily
usable to someone who doesn’t have Guix-Jupyter. That’s a limitation,
but one that’s probably hard to avoid.
Is a kernel the right approach to adding reproducible deployment to Jupyter? Should it be a built-in feature of Notebook or of Jupyter Lab? Maybe. There’s an engineering argument that Jupyter probably shouldn’t be tied to a specific deployment tool, and in that sense, handling it as a kernel or as an extension leaves Jupyter users a freedom of choice.
No matter what approach is used, our best-practices book should be updated so that Jupyter notebooks lacking deployment information become a thing of the past!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).





