Guix-HPC Activity Report, 2020
This document is also available as PDF (printable booklet).
Guix-HPC is a collaborative effort to bring reproducible software deployment to scientific workflows and high-performance computing (HPC). Guix-HPC builds upon the GNU Guix software deployment tool and aims to make it a better tool for HPC practitioners and scientists concerned with reproducible research.
Guix-HPC was launched in September 2017 as a joint software development project involving three research institutes: Inria, the Max Delbrück Center for Molecular Medicine (MDC), and the Utrecht Bioinformatics Center (UBC). GNU Guix for HPC and reproducible science has received contributions from additional individuals and organizations, including CNRS, the University of Tennessee Health Science Center (UTHSC), the Leibniz Institute for Psychology (ZPID), Cray, Inc. (now HPE), and Tourbillion Technology.
This report highlights key achievements of Guix-HPC between our previous report a year ago and today, February 2021. This year was marked by two releases (version 1.1.0 in April 2020 and version 1.2.0 in November) as well as releases of the Guix Workflow Language and of Guix-Jupyter.
Outline
Guix-HPC aims to tackle the following high-level objectives:
- Reproducible scientific workflows. Improve the GNU Guix tool set to better support reproducible scientific workflows and to simplify sharing and publication of software environments.
- Cluster usage. Streamlining Guix deployment on HPC clusters, and providing interoperability with clusters not running Guix.
- Outreach & user support. Reaching out to the HPC and scientific research communities and organizing training sessions.
The following sections detail work that has been carried out in each of these areas.
Reproducible Scientific Workflows
Supporting reproducible research in general remains a major goal for Guix-HPC. The ability to reproduce and inspect computational experiments—today’s lab notebooks—is key to establishing a rigorous scientific method. We believe that a prerequisite for this is the ability to reproduce and inspect the software environments of those experiments.
We have made further progress to ensure Guix addresses this use case. We work not only on deployment issues, but also upstream—ensuring source code is archived at Software Heritage—and downstream—devising tools and workflows for scientists to use.
The Guix Workflow Language
The Guix Workflow Language (or GWL) is a scientific computing extension to GNU Guix's declarative language for package management. It allows for the declaration of scientific workflows, which will always run in reproducible environments that GNU Guix automatically prepares. In the past year the GWL has seen further development to refine the language and its integration with the Guix environment management features.
The GWL was always intended to be used as an extension to Guix, but
this presented unique problems. Extensions to Guix may use the Guile
modules it provides as a library in addition to accessing Guix package
definitions. With the exact version of Guix under user control via
guix pull this put extension developers between a rock and a hard
place: they could either dynamically bind to the version of Guix
currently in use and hope that the API of future versions of Guix
remains unchanged compared to the version they used during
development, or they could restrict the extension to a well-known
version of Guix with the unfortunate side effect that any reference to
packages would be outdated compared to those available in the variant
of Guix currently installed by the user.
Neither of these options appealed to the developers of the Guix Workflow Language, so they implemented a third option based on the Guix API for inferiors. The result is that any package requested by users of the Guix Workflow Language will be resolved with the currently installed variant of Guix, including any defined channels or time travel settings. For all other features, the Workflow Language binds to a well-known version of Guix as a library, thereby satisfying the seemingly conflicting expectations and requirements of users and developers.
The requirements of the GWL motivated the implementation of a generic
mechanism in Guix to discover extensions, as well as performance
improvements in the inferior mechanism.
Version 0.3.0 of the Guix Workflow Language was announced in early February 2021.
Guix-Jupyter
We announced Guix-Jupyter a bit more than a year ago, with two goals: making notebooks self-contained or “deployment-aware” so that they automatically deploy the software (and data!) that they need, and making said deployment bit-reproducible.
In essence, Guix-Jupyter treats software deployment and data as a first-class input to the computation described in the notebook. One can run the notebook on one machine or another, today or two years from now, and be sure it’s running in the exact same software environment.
This Jupyter “kernel” builds on Guix support for reproducible builds and for “time travel”. That very first version demonstrated what can be achieved, and it addressed what remains a very relevant issue in the Jupyter world, as is clear to anyone who has tried to run a notebook published in one of the many public galleries.
Version 0.2.1 was announced in January
2021.
Among the user interface changes and bug fixes it provides, ;;guix describe and ;;guix search “magic” commands have been added,
providing the same functionality as the same-named guix commands.
Build and download progress is now reported in the cell that triggered
it, which improves the user experience. While we still consider it
“beta”, we believe it already smoothly covers a wide range of use cases.
From Source Code to Research Article
GWL and Guix-Jupyter are both tools designed for a scientific audience, as close as possible to the actual scientific experiments and workflows. In the same vein, we participated in the Ten Years Reproducibility Challenge organized by ReScience C, an open-access, peer-reviewed journal that targets computational research and encourages the replication of already published research.
Participants were invited to pick a scientific article they had published at least ten years earlier, and to try and reproduce its results. Needless to say, participants encountered many difficulties, most of which boil down to: finding the code, getting it to build, and getting it to run in an environment as close as possible to the original one.
This last challenge—re-deploying software—is of particular interest to Guix, which has to goal of supporting bit-reproducible deployments in time. Of course, the chosen articles were published before Guix even existed, but we thought it was a good opportunity to demonstrate how Guix will allow users to address these challenges from now on. Among the fifty participants, some chose to address deployment issues using Docker or virtual machines (VMs), and several chose Guix. The challenge and its contributions, including a discussion of software deployment issues and Guix, led to an article in Nature (Jeffrey M. Perkel, Challenge to scientists: does your ten-year-old code still run?, August 2020).
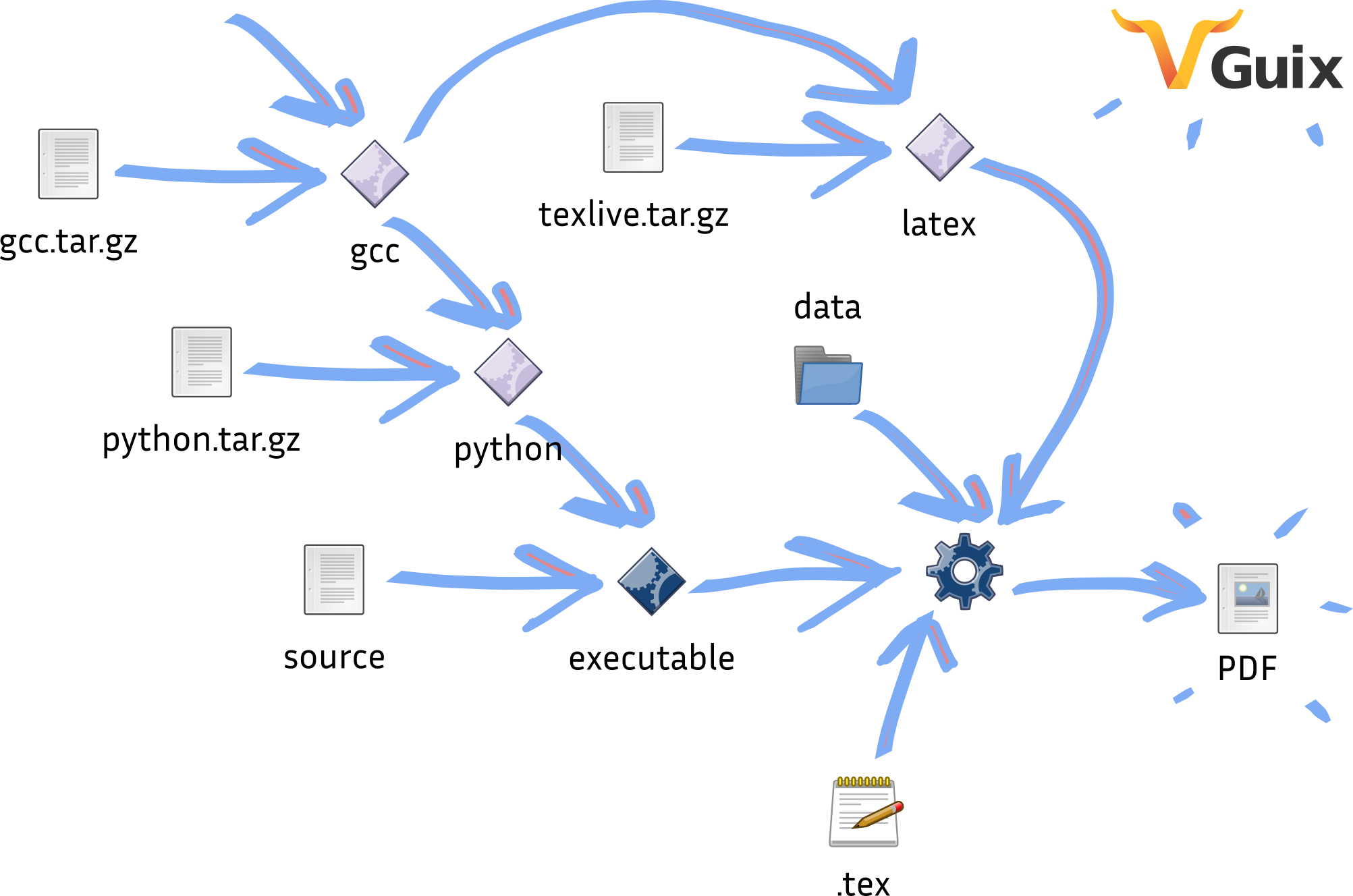
The replication work by Ludovic Courtès is an attempt to show the best one could provide: a complete, end-to-end reproducible research article pipeline, from source code to PDF. The articles shows how to bridge together code that deploys the software evaluated in the paper, scripts that run the evaluation and produce plots, and scripts that produce the final PDF file from LaTeX source and plots. The end result is approximately 400 lines of code that allow Guix to rebuild the whole article and the experiment it depends on with a well-specified, reproducible software environment.
The article concludes on our vision:
We hope our work could serve as the basis of a template for reproducible papers in the spirit of Maneage. We are aware that, in its current form, our reproducible pipeline requires a relatively high level of Guix expertise—although, to be fair, it should be compared with the wide variety of programming languages and tools conventionally used for similar purposes. We think that, with more experience, common build processes and idioms could be factorized as libraries and high-level programming constructs, making it more approachable.
[…] We look forward to a future where reproducible scientific pipelines become commonplace.
This is just the beginning. We plan to keep working closely with scientists and journals such as ReScience C to investigate ways to make this approach more widely applicable.
Soon after the challenge, we organized a one-day on-line hackathon to collectively work on providing missing bits so more scientific experiments can be reproduced. This led to improved coverage of historic package versions in the new Guix-Past channel, which was created for the challenge.
Ensuring Source Code Availability
Guix, for instance with guix time-machine, allows you to travel back in time and
to re-deploy software reproducibly. It is a great step forward in support of
reproducible research. But there is still an important piece to make this
viable: a stable source code archive. The collaboration with Software
Heritage (SWH) initiated in
2019
is continued.
Since March
2019,
Guix eases the request submission to SWH for long-term archiving via guix lint -c archival. Once the package is ready,
linting ensures the source code is saved on SWH. Later, if
original upstream source code vanishes, Guix falls back to
SWH. This aforementioned article read:
Our plan is to extend Software Heritage such that it would periodically archive the source code of software packaged by Guix.
This is now the case: Guix serves the file
sources.json, which contains
references to all the source code packages refer to; this is periodically
ingested by Software Heritage via the SWH
nixguix
“loader”. As a result, more packages are continuously archived. This
work was done by Tweag for the benefit of the Nix and Guix projects; the
Guix team helped review it.
The primary issue that remains before SWH covers all the needs of Guix
is the availability of source code “tarballs” (archives such as
tar.gz files). SWH archives the contents of version control
repositories and tarballs—not tarballs themselves. This makes
sense from an engineering viewpoint, but it means that tools such as Guix
that depend on those tarballs cannot simply fetch them by hash.
However, most Guix packages currently refer to tarballs rather than
version-control system (VCS) repositories. For these, the automatic SWH
fallback in Guix will be of little help.
Guix developer Timothy Sample started working on a solution to address this issue. Sample developed a tool called Disarchive that aims to create a link between tarballs and their contents. Concretely, Guix would maintain a Disarchive database mapping source code tarball hashes to their content hash along with tarball metadata. This would allow Guix to fetch content from SWH while still being able to reconstruct the expected tarball and check that it corresponds to the hash contained in the package definition. This work is still in a prototyping phase but is already promising.
Package Transformations
HPC practitioners are often demanding when it comes to customizing software they deploy: choosing dependencies, versions, build flags, and so on. Guix caters to these needs through its programming interfaces but also via its easy-to-use package transformation options.
Over the past year, several of them were added. The
--with-c-toolchain option allows users to build a package and all its
dependents with a given C toolchain; this is useful for those willing to
benefit from the latest improvements in optimizing compilers. The
--with-patch option applies a patch to a package and rebuilds it along
with its dependent. The --with-debug-info rebuilds a package in a way
that preserves its debugging info (if it wasn’t already available) and
without requiring a rebuild of its dependents.
Furthermore, package transformation options passed to guix install and
similar commands are now recorded in profiles and replayed when
running guix upgrade, thereby ensuring user customization is
preserved. Another novelty is that package transformations now apply to
“implicit inputs” (package dependencies provided by the build system),
giving users even more flexibility.
The Guix reference manual now includes a section discussing the various ways to define package variants.
Declarative Deployment with Manifests
In addition to the “imperative” management style where one runs guix install and related commands, Guix supports a declarative style where
the user provides a code snippet, called a manifest, that describes
the packages to deploy. The declarative approach has the advantage of
being explicit and stateless: the manifest file can be put under version
control and shared with others. It is also flexible because users have
access to the whole programming environment of Guix from within the
manifest.
To reduce the learning curve and make it easier to switch to the
declarative style, we made two improvements. First, users can now
export a manifest from an existing profile by running guix package --export-manifest. The output can be stored in a file and users can
switch to running guix package --manifest with that file right away.
Second, there is now a high-level programming interface giving access to
package transformations, with a one-to-one mapping to their command-line
syntax. This interface can be used in manifests. In fact, the manifest
produced by --export-manifest uses it when needed.
Packaging
The package collection that comes with Guix keeps growing. It now contains 16,000 packages, including many scientific packages ranging from run-time support software such as implementations of the Message Passing Interface (MPI), to linear algebra software, to statistics and bioinformatics modules for R. In addition to packages in Guix proper, we have been maintaining channels, which provide additional packages, usually specialized in one application domain.
As part of the aforementioned Ten Years Reproducibility Challenge and subsequent reproducible research hackathon, we created the Guix-Past channel, with the goal of collecting versions of software older than Guix itself, for use in reproducible research experiments. It now contains sixty packages, including Python 2.4 (from 2008), Perl 5.14 (from 2011), and 2006-era versions of the GNU “Autotools”. As an example, Guix-Past allowed Genenetwork administrators to migrate a 20-year old web service from a 12-year old CentOS and it is now running in a modern Guix container. We expect Guix-Past will serve as the basis for future reproducible research endeavors.
A new Guix Science channel has been established to collaborate on packaging and maintaining recent scientific software that currently cannot be included into Guix proper. JASP, RStudio and JupyterLab are available. A substitute server continuously builds these packages and serves binary substitutes for them.
Since Guix is trivially extensible, users do not need to wait for the
community to prepare missing packages they may need right now. Tools
like the new package importer and installer for
R let users of
the R programming language install any R package—from CRAN,
Bioconductor, or any Mercurial or Git repository—by leveraging the
recursive package importing facilities that Guix provides from within
a running R session. This replaces the need for installers like
devtools and exposes reproducible package management features
through a familiar interface within R.
Cluster Usage and Deployment
Over the last few years, we have been developing guix pack to support
users who target supercomputers where Guix is unavailable. guix pack
creates “application bundles” as Docker or Singularity images, or as
plain self-contained tarballs. We previously implemented relocatable
packs: archives that can be extracted anywhere on the file system while
still allowing users to run the software therein. The appeal of this
option is that it allows users to run packaged software on machines that
provide neither Guix nor a container run-time.
Relocatable packs were initially implemented by using the Linux
“unprivileged user namespace” feature, with a fallback to PRoot when
unprivileged user namespaces are unavailable. In the latter case
though, the overhead can be prohibitively high for some applications.
To address that, guix pack can now build faster relocatable
packs
that rely on a customization of the run-time linker (ld.so) along with
the use of Fakechroot. It provides the flexibility of packs with
negligible overhead.
Guix deployment on scientific clusters continues. A notable example is work-in-progress to support Guix out-of-the-box on Grid’5000, a major French grid and HPC environment. Installing Guix on Grid’5000 is quite different from installing it on a “regular” HPC cluster due to the unusual nature of this environment: Grid’5000 users have the option to deploy their own operating system images on compute nodes and have administration privileges on them. This has required adjustments compared to our cluster installation guide. This should be in production in the coming weeks.
This fall at UTHSC, Memphis TN, we have installed an 11-node HPC Octopus cluster (264 cores) for pangenome and genetics research. Notable about this HPC is that it is administered by the users, thanks to GNU Guix: we install, run and manage the cluster as researchers. UTHSC IT manages the infrastructure, i.e., physical placement, routers and firewalls, but beyond that there are no demands on IT. Thanks to out-of-band access we can completely (re)install machines remotely. Octopus runs GNU Guix on top of a minimal Debian install and we are experimenting with pure GNU Guix nodes that can be run on demand. Lizardfs is used for distributed network storage. Almost all deployed software has been packaged in GNU Guix and can be installed by regular users on the cluster without root access.
Guix is also the foundation for the data analysis platform PsychNotebook, which assists researchers and students in the preparation, analysis, and transparent documentation of psychological data. While a modern web interface hides most aspects of Guix from the user, under the hood channel files and manifests are used to create shareable and reproducible environments, which then can be shared with other users of the platform or exported and run on local hardware. After a testing period in the previous semester with about 15 students the system is now live and currently used to teach about 20 students the basics of R using RStudio Web.
Outreach and User Support

Guix-HPC is in part about “spreading the word” about our approach to reproducible software environments and how it can help further the goals of reproducible research and high-performance computing development. This section summarizes articles, talks, and training sessions given this year.
Articles
The following articles were published in the Ten Years Reproducibility Challenge special issue of the ReScience C journal (volume 6, issue 1); they present different ways of using Guix to reproduce the software environment associated with a past computational experiment:
- Andreas Enge, [Re] Volume computation for polytopes: Vingt ans après
- Konrad Hinsen, [Rp] Structural flexibility in proteins — impact of the crystal environment
- Ludovic Courtès, [Re] Storage Tradeoffs in a Collaborative Backup Service for Mobile Devices
Hinsen also wrote about “staged computations”, their use as part of reproducible research workflows, and how Guix can help:
- Konrad Hinsen, Staged computation: the technique you didn't know you were using, Computing in Science and Engineering (CISE), 22 (4)
We published 8 articles on the Guix-HPC blog touching topics such as fast relocatable packs, reproducible research article workflows, and Jupyter integration.
Talks
Since last year, we gave the following talks at the following venues:
- JDEV conference (on-line), July 2020 (Ludovic Courtès)
- on-line seminar of the Belgium Research Software Engineers community (BE-RSE), Nov. 2020 (Ludovic Courtès)
- on-line Guix Days, Nov. 2020 (Lars-Dominik Braun)
- FOSDEM HPC track, Feb. 2021 (Ricardo Wurmus)
We also organised the following events:
- an online reproducible research hackathon, where 15 people were connected to tackle issues inspired by contributions from the Ten Years Reproducibility Challenge organized by ReScience;
- the first online GNU Guix Day, which attracted more than 50 people all around the world;
- GNU Guix Days
Training Sessions
JDEV, the annual conference gather research engineers from all the French research institutes and universities, took place on-line in July 2020. It included an presentation of Guix along with an introductory workshop.
The User Tools for HPC (UST4HPC) workshop took place in January 2021. It is organized as part of the training sessions program of the French national scientific research center (CNRS). It included talks and hands-on session about Guix and Guix-Jupyter.
Personnel
GNU Guix is a collaborative effort, receiving contributions from more than 60 people every month—a 50% increase compared to last year. As part of Guix-HPC, participating institutions have dedicated work hours to the project, which we summarize here.
- CNRS: 0.25 person-year (Konrad Hinsen)
- Inria: 2 person-years (Ludovic Courtès, Maurice Brémond, and the contributors to the Guix-HPC channel: Emmanuel Agullo, Florent Pruvost, Gilles Marait, Nathalie Furmento, Marek Felšöci, Adrien Guilbaud, Philippe Swartvagher, Matthieu Simonin)
- Max Delbrück Center for Molecular Medicine (MDC): 2 person-years (Ricardo Wurmus and Mădălin Ionel Patrașcu)
- Tourbillion Technology: 0.8 person-year (Paul Garlick)
- Université de Paris: 0.5 person-year (Simon Tournier)
- University of Tennessee Health Science Center (UTHSC): 1.0 person-year (Efraim Flashner, Bonface Munyoki, Erik Garrison and Pjotr Prins)
- Utrecht Bioinformatics Center (UBC): 0.1 person-year (Roel Janssen)
Perspectives
Guix availability on scientific computing clusters remains a top
priority. We have seen increased adoption and interest among cluster
system administrators. The latest guix pack improvements should be
welcomed by those targeting clusters not running Guix. We expect to
continue work on these two complementary fronts: reaching out to system
administrators, notably through training sessions, and streamlining the
use of packs and non-root deployment in general.
Upstream, we will continue to work with Software Heritage with the goal of achieving complete archive coverage of the source code Guix refers to. We have identified challenges related to source code “tarball” availability; this will probably be one of the main efforts in this area for the coming year.
Downstream, a lot of work has happened in the area of reproducible research tools. First, our package collections have grown to include more and more scientific tools; the new Guix Science and Guix-Past channels cater to additional use cases. Second, the Guix Workflow Language, Guix-Jupyter, have matured; along with the PsychNotebook service, they bridge the gap between reproducible software deployment and reproducible scientific tools and workflows. Likewise, our participation in the “Ten Years Reproducibility Challenge” demonstrated very concretely what Guix brings to scientific workflows. It showed that fully reproducible research article workflows are indeed a possibility.
Working on the tools and workflows directly in the hands of scientists will be a major focus of the coming year. We want to contribute to raising the bar of what scientists come to expect in terms of reproducible workflows.
There’s a lot we can do and we’d love to hear your ideas!
Unless otherwise stated, blog posts on this site are copyrighted by their respective authors and published under the terms of the CC-BY-SA 4.0 license and those of the GNU Free Documentation License (version 1.3 or later, with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts).






